Abstract
Background
Mutations in the NK2 homeobox 1 (NKX2-1) gene are associated with lung disease in infants and children. We hypothesize that disruption of normal surfactant gene expression with these mutations contributes to the respiratory phenotypes observed.
Methods
To assess transactivational activity, cotransfection of luciferase reporter vectors containing surfactant protein B or C (SFTPB or SFTPC) promoters with NKX2-1 plasmids was performed and luciferase activity was measured. To assess the binding of mutated proteins to target DNA, electrophoretic mobility shift assays (EMSA) were performed using nuclear protein labeled with oligonucleotide probes representing NKX2-1 consensus binding sequences followed by gel electrophoresis. The effect of overexpression of wild-type (WT) and mutant NKX2-1 on SFTPB and SFTPC was evaluated with quantitative real-time PCR.
Results
Decreased transactivation of the SFTPB promoter by both mutants and decreased transactivation of the SFTPC promoter by the L197P mutation was observed. EMSA demonstrated decreased DNA binding of both mutations to NKX2-1 consensus binding sequences. Transfection of A549 cells with NKX2-1 expression vectors demonstrated decreased stimulation of SFTPB and SFTPC expression by mutant proteins compared with that of WT.
Conclusion
Disruption of transcriptional activation of surfactant protein genes by these DNA-binding domain mutations is a plausible biological mechanism for disruption of surfactant function and subsequent respiratory distress.
Similar content being viewed by others
The NK2 homeobox 1 (NKX2-1) gene encodes thyroid transcription factor-1 (TTF-1), a critical regulator of gene transcription in the brain, thyroid gland, and lung (1,2). The 4 kb NKX2-1 is located on human chromosome 14 (3,4) and includes a DNA-binding homeodomain (HD) that encodes amino acids 189–253 (Figure 1). Mutations in this region of the gene disrupt transcriptional activity of TTF-1 target genes by altering the binding of TTF-1 to elements within target genes (5,6,7,8). NKX2-1 expression is essential for early lung morphogenesis, as well as later regulation of the expression of genes necessary for lung function, including the surfactant genes SFTPB or SFTPC (5,9,10,11,12). The molecular mechanisms by which NKX2-1 mutations result in lung disease phenotypes remain poorly understood.
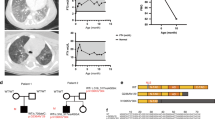
NK2 homeobox 1 (NKX2-1) gene showing three exons and the region encoding the homeodomain (HD) located in exon 3. The two patients described in the manuscript had mutations in the homeodomain at nucleotide 583 (c.583C>T) and nucleotide 590 (c.590T>C), resulting in the R195W and L197P amino-acid substitutions, respectively.
The incidence of respiratory distress associated with mutations in the NKX2-1 gene is unknown. Pulmonary phenotypes observed in infants and children with brain–thyroid–lung syndrome and NKX2-1 mutations are diverse, including respiratory distress in the newborn period, interstitial lung disease in infancy or childhood, and recurrent respiratory infections (3,10,13,14,15,16,17,18,19,20). Affected infants and children may also demonstrate neurological abnormalities such as newborn hypotonia, developmental delay in infancy, and/or movement disorders in childhood, as well as hypothyroidism (21,22,23,24,25,26,27).
We previously described 21 individuals with brain–thyroid–lung syndrome, 16 of whom presented with neonatal respiratory distress (10). Two of these individuals presented with respiratory distress syndrome and progressed to severe interstitial lung disease that ultimately required lung transplantation. These individuals were found to have heterozygous, de novo, missense mutations in the NKX2-1 HD (R195W and L197P). Neither child had mutations identified in SFTPB or SFTPC (10). The HD location of these mutations prompted us to investigate the encoded transcriptional mechanisms that disrupt surfactant gene expression in these patients.
Previous studies, including ours with these same patients, have noted the presence of surfactant proteins in lung samples and in lung lavage (3,10,19,28). In some cases, there appears to be decreased expression of surfactant proteins in the presence of NKX2-1 mutations, and in other cases, surfactant protein expression appears to be unchanged or possibly increased. These results, however, are difficult to interpret in the context of various treatments, including mechanical ventilation, oxygen, and medications to which these patients have been exposed. It becomes difficult to separate the changes in gene expression directly due to the mutations from the effects of these treatments. Therefore, we turned to an in vitro system to demonstrate the biological plausibility that these mutations are disease causing.
Methods
Patient Selection
Subjects were identified retrospectively through protocols approved by the Washington University School of Medicine Institutional Review Board by sequencing SFTPB, SFTPC, and NKX2-1 in a cohort of infants who had undergone lung transplantation for respiratory failure (10). Neither patient had an identified etiology or pathogenic mutations in SFTPB or SFTPC (10). Written informed consent was obtained from the parents of each patient.
Cell Culture
The human epithelial cell line, A549, was cultured in F12K medium with 10% fetal bovine serum and penicillin/streptomycin (standard medium) at 37 °C. In some experiments, SFTPC expression was induced with 33 nM dexamethasone (29).
Site-Directed Mutagenesis
The mutations c.583C>T (R195W) and c.590T>C (L197P) were introduced by site-directed mutagenesis (Stratagene, La Jolla, CA) into the pRC/CMV/NKX2-1 expression vector that contained the full-length, wild-type, human NKX2-1 cDNA. Construct sequences were confirmed by Sanger sequencing. Forward primer 5′--3′ and reverse primer 5′--3′ were used to introduce the R195W mutation. Forward primer 5′--3′ and reverse primer 5′--3′ were used for the L197P mutation. NKX2-1 encodes two isoforms that differ by 30 amino acids (11). For this study, numbering of residues based on the longer, 401-amino-acid isoform was used to be consistent with our previous report and Human Genome Variation Society nomenclature (10). Other authors have used numbering based on the shorter isoform (3). The wild-type NKX2-1 plasmid was a kind gift from Parviz Minoo (University of Southern California, Los Angeles, CA).
Transient Transfection
The SFTPB promoter or SFTPC promoter was cloned into a firefly luciferase vector lacking a promoter (pGL4.10[luc2]; Promega, Madison, WI) to create reporter plasmids (SFTPB_luc and SFTPC_luc). The SFTPB promoter was a 1.0 kb human promoter and the SFTPC promoter was a 3.7 kb human promoter (kind gift from Dr J. Whitsett, Cincinnati Children’s Hospital, Cincinnati, OH). A549 cells (1.5 × 105 cells/well in 12-well plates 24 h before transfection) were cotransfected using Fugene 6 (Roche Applied Science, Indianapolis, IN) and 0.1 μg of either the SFTPB_luc or SFTPC_luc reporter plasmid together with 0.1 μg of pRC/CMV expressing wild-type or mutant NKX2-1. Cotransfection with a Renilla-luciferase expression vector (pRL-TK) was used to normalize for cell transfection efficiency. Transfections with empty pRC/CMV served as a control. After 48 h, cells were lysed, and firefly luciferase and then Renilla luciferase were measured in a sequential manner with a dual luciferase assay (Promega). After normalization of firefly luciferase activity with Renilla luciferase activity, normalized luciferase activities were compared between the wild-type NKX2-1 plasmids and the mutations using Student’s paired t tests.
Electrophoretic Mobility Shift Assay
To assess the ability of the mutated TTF-1 proteins to bind to target DNA sequences in the promoter region of SFTPB or SFTPC, electrophoretic mobility shift assays were performed. A549 cells (4 × 106/well) were transfected with 8 μg of wild-type or mutant NKX2-1 expression vectors by nucleofection (Lonza, Basel, Switzerland) according to the manufacturer’s instructions. Nuclear protein was isolated 48 h after transfection and extracted using nuclear and cytoplasmic extraction reagents (NE-PER; Thermoscientific, Rockford, IL). Next, 6 μg of nuclear protein was incubated with IRDye 700-labeled DNA probes (Integrated DNA Technologies, Coralville, IA) that included SFTPB- or SFTPC-specific TTF-1 binding sequences. Forward and reverse polyacrylamide gel electrophoresis-purified, IRDye 700-labeled probes for SFTPB were 5′--3′ and 5′--3′, respectively, and for SFTPC, 5′--3′ and 5′--3′, respectively. Electrophoretic mobility shift assay was performed using the Lightshift Chemiluminescent EMSA Kit and protocol (Thermoscientific). In some reactions, an excess of unlabeled probe (10 pmol) was added to demonstrate reaction specificity. To confirm the presence of TTF-1 protein in the reaction products, 0.8 μg of mouse anti-TTF-1 antibody (Seven Hills Bioreagents, Cincinnati, OH) was added to some reactions. The reactions were then resolved on 6% DNA acrylamide gels (Life Technologies, Grand Island, NY) and imaged with Odyssey infrared imaging system (LI-COR, Lincoln, NE).
Quantitative PCR
To assess the ability of wild-type and mutant NKX2-1 to regulate the expression of endogenously expressed SFTPB and SFTPC, quantitative real-time PCR was used in A549 cells transfected with vectors expressing wild-type or mutant NKX2-1. A549 cells (3 × 106/well) were transfected with 2 μg of wild-type or mutant NKX2-1 pRC/CMV expression vectors using Fugene 6 (Promega) according to the manufacturer’s instructions and cultured in standard media for 72 h. Because endogenous expression of SFTPC was low in A549 cells, expression was stimulated by culturing A549 cells with 33 nM dexamethasone beginning 24 h after transfection. RNA was isolated 72 h after transfection and extracted using Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Taqman Gene Expression Assays were performed in triplicate to assess gene expression of SFTPB and SFTPC in transfected vs. untransfected A549 cells.
Results
Clinical Description
Patient 1 (R195W mutation) was a male of Hispanic descent born at 38 weeks of gestation with respiratory failure at birth and treated with mechanical ventilation. He received multiple doses of surfactant replacement therapy and was discharged home on supplemental oxygen at 80 days of life. During the neonatal period, he was also diagnosed with hypothyroidism that was treated with levothyroxine. He underwent gastrostomy tube placement and Nissen fundoplication for poor feeding and concerns for aspiration. For the first 4 months of life, he remained on supplemental oxygen with multiple hospital admissions for respiratory decompensations. Computed tomography scan of his lungs showed a diffusely reticular nodular pattern. Histology from a lung biopsy showed diffuse interstitial lung disease with prominent pneumocyte hyperplasia and alveolar septal widening. Electron microscopy revealed hyperplastic type II pneumocytes with lamellar bodies of variable size that appeared crinkled and densely packed forming irregular concentric whorls. Immunohistochemistry demonstrated the presence of surfactant protein B and surfactant protein C in the tissue from his lungs, which was removed at the time of transplantation (10). Due to progressive respiratory failure, he required mechanical ventilation at 5 months of age until he underwent lung transplantation at 8 months of age. He expired at 3 years of age of pulmonary vein stenosis and pulmonary hypertension. No mutations were identified in SFTPB or SFTPC. Sequence analysis revealed a c.583C>T missense mutation in the NKX2-1 gene.
Patient 2 (L197P mutation) was a male of European descent born at 37 weeks who had respiratory distress syndrome at birth for which he was intubated, received surfactant replacement therapy, and was supported with mechanical ventilation for 12 days. His chest radiographs demonstrated a ground glass appearance. He was discharged on supplemental oxygen. At 3 months of age, he underwent fundoplication, pyloroplasty, and gastrostomy tube placement for poor oral feeding and possible aspiration. At 5 months of age, he was hospitalized for respiratory syncytial virus and received systemic steroids. Computed tomography scan of his lungs showed generalized ground glass opacities and lung hyperinflation. Histology from lung biopsy showed nonspecific parenchymal changes, including diffuse pneumocyte hyperplasia, mild to focally moderate interstitial widening, and increased numbers of histocytic-like cells in the alveolar septae. Histology also demonstrated mild inflammatory infiltrates, including lymphocytes, scattered plasma cells, eosinophils, and intra-alveolar foamy macrophages. Electron micrographs revealed numerous lamellar bodies of various sizes and some with a “fried egg” appearance (10). Immunohistochemistry demonstrated the presence of surfactant protein B and surfactant protein C in the tissue from his lungs, which was removed at the time of transplantation (10). Owing to progressive respiratory failure and interstitial lung disease, he underwent lung transplantation at 10 months of age. He developed attention-deficit/hyperactivity disorder at 10 years of age and a choreiform movement disorder at 12 years of age. Thyroid studies were normal. He is now 17 years old and has maintained his first set of donor lungs, although his course is now complicated by bronchiolitis obliterans. No mutations were identified in SFTPB or SFTPC. Sequence analysis revealed a c.590T>C missense mutation in the NKX2-1 gene.
Transient Transfection
To investigate disruption of transcriptional activity encoded by the R195W and L197P mutants, transient cotransfection was performed with NKX2-1 wild-type or mutant expression vectors and SFTPB or SFTPC luciferase reporter vectors. Cotransfection of a wild-type NKX2-1 expression vector with an SFTPB-luciferase reporter increased luciferase activity sevenfold compared with that of the vector alone (luc2) (P<0.05) (Figure 2a). Cotransfection with the R195W or L197P mutation expression vectors decreased SFTPB promoter luciferase activity by 34% and 50%, respectively, compared with that of wild-type NKX2-1 (P<0.05 for each) (Figure 2a).
Transactivation of surfactant protein B (SFTPB) (a) and surfactant protein C (SFTPC) (b) promoters by wild-type and mutant NK2 homeobox 1 (NKX2-1). Relative luciferase activity is indicated following correction for transfection efficiency. Error bars indicate standard error of the mean. *P<0.05 vs. vector alone. **P<0.05 vs. wild type.
Cotransfection of the wild-type NKX2-1 expression vector with the SFTPC reporter construct increased luciferase activity 15-fold compared with that of the vector alone (luc2) (P<0.05) (Figure 2b). Cotransfection with the R195W mutation expression vector did not change luciferase activity, whereas cotransfection with the L197P mutation expression vector decreased luciferase activity of the SFTPC promoter by 25% compared with that of the wild-type vector (P<0.05) (Figure 2b).
Electrophoretic Mobility Shift Assay
To evaluate the binding of mutant TTF-1 proteins to SFTPB- or SFTPC-specific NKX2-1 target promoter sequences, electrophoretic mobility shift assays were performed with nuclear protein extracted from transiently transfected A549 cells. Nuclear protein from A549 cells transfected with wild-type TTF-1 was bound to the SFTPB target promoter sequence (Figure 3a; lane 2), and binding was inhibited by excess unlabeled probes (Figure 3a; lane 3). When quantified with infrared imaging and compared with wild-type TTF-1 protein, nuclear protein from A549 cells transfected with the R195W or L197P mutant decreased binding to SFTPB target promoter sequence by 33% (Figure 3a; lane 4) and 15%, respectively (Figure 3a; lane 6). The binding reactions were inhibited by excess unlabeled probes (Figure 3a; lanes 5 and 7). Supershift (arrow) with anti-TTF-1 antibody indicated the presence of TTF-1 protein in the protein–DNA complex (Figure 3a; lane 8). Each experiment was repeated a minimum of three times with consistent changes observed in binding.
Electrophoretic mobility shift assays were performed with nuclear protein, including wild-type or mutant thyroid transcription factor-1 (TTF-1). Reactions were performed with labeled surfactant protein B (SFTPB) (a) and surfactant protein C (SFTPC) (b) TTF-1 binding sites. In some reactions, unlabeled probe or anti-TTF-1 antibody was included. The white arrow indicates the TTF-1 binding complex; the dark arrow indicates the supershifted binding complex.
Wild-type TTF-1 was also bound to the SFTPC-specific target promoter sequence (Figure 3b; lane 2), and binding was inhibited by excess unlabeled probe (Figure 3b; lane 3). Nuclear protein from A549 cells transfected with each mutation decreased binding by 30% compared with that of wild-type TTF-1 (Figure 3b; lanes 4 and 6). Binding of both mutant proteins was inhibited by the addition of excess unlabeled probe (Figure 3b; lanes 5 and 7). Supershift (arrow) with anti-TTF-1 demonstrates the presence of TTF-1 protein in the protein–DNA complex (Figure 3b, lane 8). Each experiment was repeated a minimum of three times with consistent changes in binding observed.
Endogenous SFTPB and SFTPC Expression after Transfection with Wild-Type or Mutant NKX2-1 Vectors
To examine the effect of wild-type and mutant TTF-1 on the expression of surfactant protein genes, vectors expressing wild-type or one of the mutant NKX2-1 genes were transfected into A549 cells. SFTPB and SFTPC transcript expression was then measured by quantitative PCR. Transfection of A549 cells with a wild-type NKX2-1 expression vector increased endogenous SFTPB expression 64-fold (P<0.05) (Figure 4a). Transfection of the R195W or L197P NKX2-1 mutants decreased endogenous SFTPB expression by 63% and 95%, respectively, compared with that of wild-type (P<0.05) (Figure 4a).
Expression of surfactant protein B (SFTPB) (a) and surfactant protein C (SFTPC) (b) by A549 cells in response to wild-type or mutant NK2 homeobox 1 (NKX2-1) expression. Relative transcript abundance is shown compared with that of untransfected cells. Error bars indicate standard error of the mean. *P<0.05 vs. untransfected cells. **P<0.05 vs. cells transfected with wild-type vector.
Because endogenous expression of SFTPC by A549 cells is low, A549 cells were cultured in the presence of dexamethasone to increase SFTPC expression. Transfection of A549 cells cultured in the presence of dexamethasone with wild-type NKX2-1 increased the endogenous SFTPC expression 9.0-fold (P<0.05) (Figure 4b). Transfection of the R195W or L197P NKX2-1 mutants decreased endogenous SFTPC expression by 73% and 76%, respectively (P<0.05) (Figure 4b).
Discussion
Advances in computational prediction of the functional impact of DNA sequence changes on gene function have enabled the identification of genotype–phenotype associations. However, confirmation of pathogenicity requires model systems to demonstrate functional disruption of computationally discovered genomic variants. The NKX2-1 HD mutations described here were identified in patients with extreme respiratory phenotypes who required lung transplantation. Both patients had brain–thyroid–lung syndrome, although some of the neurologic symptoms in patient 1 may have been confounded by his illness severity (10). The phenotypes of patient 1 and a previously reported patient with the same mutation are similar (3). The mutation in patient 2 (L197P) is novel.
Functional analysis of these mutations suggests that both of them encode TTF-1 mutant proteins with decreased binding to NKX-2-specific target sites in the SFTPB and SFTPC promoters. Both mutants also demonstrated reduced upregulation of endogenous SFTPB and SFTPC expression compared with that of wild-type mutants. Both mutants resulted in reduced SFTPB promoter activation, but only the L197P mutant reduced SFTPC promoter activation with no significant effect of the R195W mutant. The absence of an effect of the R195W mutation on SFTPC promoter transactivation, despite decreased binding of the mutant protein, may be the result of the unique characteristics of the truncated promoter used for transient transfection vs. the endogenous promoter. These results are also in contrast to the results of Guillot et al. (3) where increased activation of the SFTPC promoter was reported following cotransfection with the R195W mutant (R165W in their manuscript). This difference may be attributable to the different SFTPC promoters tested. We used a 3.7 kb human SFTPC promoter, whereas Guillot et al. (3) used a 4.8 kb mouse SFTPC promoter in their studies.
The genotype–phenotype association of the NKX2-1 mutations in our patients and those of other patients with NKX2-1 disease-associated mutations is consistent (10,18,27,30,31) and suggests loss-of-function haploinsufficiency or gain-of-function dominant-negative mechanisms. In addition to disruption of the TTF-1 DNA-binding HD encoded by the mutations in our two patients, NKX2-1 mutations in two transactivation domains, a nuclear localization signal domain and functionally important phosphorylation sites (32,33,34,35), have also been described (3,10,14,18,22,26,27,31,36,37,38,39).
Disruption of the transactivating interaction of TTF-1 with the surfactant-associated genes suggests disrupted surfactant composition or function as a mechanism by which mutations in NKX2-1 result in a respiratory phenotype. Although surfactant proteins are present in the lungs of the patients reported here at the time of transplantation in what appears to be normal or even increased amounts, it is difficult to interpret these data in the context of the multiple treatments these patients have received over a period of months. The use of animal models would be helpful in distinguishing between the direct effects of the mutations and the effect of treatments on surfactant protein expression in vivo. In addition to its role in surfactant metabolism, NKX2-1 also has a critical role in early lung development, as well as interacting with a large number of other genes in addition to the surfactant protein genes. The targeted mutation of the mouse nkx2-1 gene resulted in a disruption of distal lung development (40). In addition, genome-wide analysis in mice revealed that TTF-1 binds to more than 1,300 genes. This included genes involved in early and late lung development, cell cycle regulation, ion transport, RAS signal transduction, and response to injury (7). Thus, these other fundamental disturbances in lung development and function could contribute to the respiratory phenotype seen in these patients. There is suggested disruption of normal lung growth in these patients but, once again, it is difficult to separate the effects of the mutation from the effects of treatments (10). This again underscores the importance of in vitro surrogate cell systems and animal models in unraveling the pathogenic mechanisms involved. Careful phenotyping of patients who are identified with NKX2-1 mutations will provide opportunities for further characterization of the mechanisms of respiratory dysfunction.
References
Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 1991;113:1093–1104.
Trueba SS, Auge J, Mattei G et al. PAX8, TITF1, and FOXE1 gene expression patterns during human development: new insights into human thyroid development and thyroid dysgenesis-associated malformations. J Clin Endocrinol Metab 2005;90:455–462.
Guillot L, Carre A, Szinnai G et al. NKX2-1 mutations leading to surfactant protein promoter dysregulation cause interstitial lung disease in "Brain-Lung-Thyroid Syndrome". Hum Mutat 2010;31:E1146–E1162.
Hamdan H, Liu H, Li C et al. Structure of the human Nkx2.1 gene. Biochim Biophys Acta 1998;1396:336–348.
Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin Sci (Lond) 2009;116:27–35.
Provenzano C, Zamboni M, Veneziano L et al. Functional characterization of two novel mutations in TTF-1/NKX2.1 homeodomain in patients with benign hereditary chorea. J Neurol Sci 2016;360:78–83.
Tagne JB, Gupta S, Gower AC et al. Genome-wide analyses of Nkx2-1 binding to transcriptional target genes uncover novel regulatory patterns conserved in lung development and tumors. PLoS ONE 2012;7:e29907.
Thorwarth A, Schnittert-Hubener S, Schrumpf P et al. Comprehensive genotyping and clinical characterisation reveal 27 novel NKX2-1 mutations and expand the phenotypic spectrum. J Med Genet 2014;51:375–387.
Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem 1996;271:6881–6888.
Hamvas A, Deterding RR, Wert SE et al. Heterogeneous pulmonary phenotypes associated with mutations in the thyroid transcription factor gene NKX2-1. Chest 2013;144:794–804.
Kolla V, Gonzales LW, Gonzales J et al. Thyroid transcription factor in differentiating type II cells: regulation, isoforms, and target genes. Am J Respir Cell Mol Biol 2007;36:213–225.
Zhou B, Zhong Q, Minoo P et al. Foxp2 inhibits Nkx2.1-mediated transcription of SP-C via interactions with the Nkx2.1 homeodomain. Am J Respir Cell Mol Biol 2008;38:750–758.
Devriendt K, Vanhole C, Matthijs G, de Zegher F. Deletion of thyroid transcription factor-1 gene in an infant with neonatal thyroid dysfunction and respiratory failure. N Engl J Med 1998;338:1317–1318.
Krude H, Schutz B, Biebermann H et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J Clin Invest 2002;109:475–480.
Pohlenz J, Dumitrescu A, Zundel D et al. Partial deficiency of thyroid transcription factor 1 produces predominantly neurological defects in humans and mice. J Clin Invest 2002;109:469–473.
Doyle DA, Gonzalez I, Thomas B, Scavina M. Autosomal dominant transmission of congenital hypothyroidism, neonatal respiratory distress, and ataxia caused by a mutation of NKX2-1. J Pediatr 2004;145:190–193.
Willemsen MA, Breedveld GJ, Wouda S et al. Brain–thyroid–lung syndrome: a patient with a severe multi-system disorder due to a de novo mutation in the thyroid transcription factor 1 gene. Eur J Pediatr 2005;164:28–30.
Carre A, Szinnai G, Castanet M et al. Five new TTF1/NKX2.1 mutations in brain-lung-thyroid syndrome: rescue by PAX8 synergism in one case. Hum Mol Genet 2009;18:2266–2276.
Kleinlein B, Griese M, Liebisch G et al. Fatal neonatal respiratory failure in an infant with congenital hypothyroidism due to haploinsufficiency of the NKX2-1 gene: alteration of pulmonary surfactant homeostasis. Arch Dis Child Fetal Neonatal Ed 2011;96:F453–F456.
Maquet E, Costagliola S, Parma J et al. Lethal respiratory failure and mild primary hypothyroidism in a term girl with a de novo heterozygous mutation in the TITF1/NKX2.1 gene. J Clin Endocrinol Metab 2009;94:197–203.
Barnett CP, Mencel JJ, Gecz J et al. Choreoathetosis, congenital hypothyroidism and neonatal respiratory distress syndrome with intact NKX2-1. Am J Med Genet A 2012;158A:3168–3173.
de Filippis T, Marelli F, Vigone MC, Di Frenna M, Weber G, Persani L. Novel NKX2-1 frameshift mutations in patients with atypical phenotypes of the brain–lung–thyroid syndrome. Eur Thyroid J 2014;3:227–233.
Ferrara JM, Adam OR, Kirwin SM et al. Brain–lung–thyroid disease: clinical features of a kindred with a novel thyroid transcription factor 1 mutation. J Child Neurol 2012;27:68–73.
Inzelberg R, Weinberger M, Gak E. Benign hereditary chorea: an update. Parkinsonism Relat Disord 2011;17:301–307.
Shetty VB, Kiraly-Borri C, Lamont P, Bikker H, Choong CS. NKX2-1 mutations in brain–lung–thyroid syndrome: a case series of four patients. J Pediatr Endocrinol Metab 2014;27:373–378.
Williamson S, Kirkpatrick M, Greene S, Goudie D. A novel mutation of NKX2-1 affecting 2 generations with hypothyroidism and choreoathetosis: part of the spectrum of brain–thyroid–lung syndrome. J Child Neurol 2014;29:666–669.
Veneziano L, Parkinson MH, Mantuano E, Frontali M, Bhatia KP, Giunti P. A novel de novo mutation of the TITF1/NKX2-1 gene causing ataxia, benign hereditary chorea, hypothyroidism and a pituitary mass in a UK family and review of the literature. Cerebellum 2014;13:588–595.
Griese M, Lorenz E, Hengst M et al. Surfactant proteins in pediatric interstitial lung disease. Pediatr Res 2016;79:34–41.
Rucka Z, Vanhara P, Koutna I et al. Differential effects of insulin and dexamethasone on pulmonary surfactant-associated genes and proteins in A549 and H441 cells and lung tissue. Int J Mol Med 2013;32:211–218.
Gras D, Jonard L, Roze E et al. Benign hereditary chorea: phenotype, prognosis, therapeutic outcome and long term follow-up in a large series with new mutations in the TITF1/NKX2-1 gene. J Neurol Neurosurg Psychiatry 2012;83:956–962.
Nettore IC, Mirra P, Ferrara AM et al. Identification and functional characterization of a novel mutation in the NKX2-1 gene: comparison with the data in the literature. Thyroid 2013;23:675–682.
De Felice M, Damante G, Zannini M, Francis-Lang H, Di Lauro R. Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J Biol Chem 1995;270:26649–26656.
Christophe-Hobertus C, Christophe D. Two binding sites for thyroid transcription factor 1 (TTF-1) determine the activity of the bovine thyroglobulin gene upstream enhancer element. Mol Cell Endocrinol 1999;149:79–84.
Ghaffari M, Zeng X, Whitsett JA, Yan C. Nuclear localization domain of thyroid transcription factor-1 in respiratory epithelial cells. Biochem J 1997;328 (Part 3): 757–761.
Silberschmidt D, Rodriguez-Mallon A, Mithboakar P et al. In vivo role of different domains and of phosphorylation in the transcription factor Nkx2-1. BMC. Dev Biol 2011;11:9.
Moya CM, Perez de Nanclares G, Castano L et al. Functional study of a novel single deletion in the TITF1/NKX2.1 homeobox gene that produces congenital hypothyroidism and benign chorea but not pulmonary distress. J Clin Endocrinol Metab 2006;91:1832–1841.
Provenzano C, Veneziano L, Appleton R, Frontali M, Civitareale D. Functional characterization of a novel mutation in TITF-1 in a patient with benign hereditary chorea. J Neurol Sci 2008;264:56–62.
Rosati A, Berti B, Melani F, Cellini E, Procopio E, Guerrini R. Recurrent drop attacks in early childhood as presenting symptom of benign hereditary chorea caused by TITF1 gene mutations. Dev Med Child Neurol 2015;57:777–779.
Salerno T, Peca D, Menchini L et al. Respiratory insufficiency in a newborn with congenital hypothyroidism due to a new mutation of TTF-1/NKX2.1 gene. Pediatr Pulmonol 2014;49:E42–E44.
Kimura S, Hara Y, Pineau T et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1996;10:60–69.
Acknowledgments
We thank Jeffrey J. Bednarski, MD of Washington University in St Louis, MO for his technical assistance with the electrophoretic mobility shift assays, Parviz Minoo, MD for the NKX2-1 wild-type cDNA, and Jeffrey A.Whitsett, MD of Cincinnati Children’s Hospital Medical Center for the human SFTPC promoter plasmid.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Statement of Financial Support
This work was supported by the National Institutes of Health (R01 HL065174 and R01 HL082747 (to FSC, AH), R21/R33 HL120760 (to FSC)), the Children’s Discovery Institute (to FSC, AH), and the Saigh Foundation (to FSC).
Rights and permissions
About this article
Cite this article
Attarian, S.J., Leibel, S.L., Yang, P. et al. Mutations in the thyroid transcription factor gene NKX2-1 result in decreased expression of SFTPB and SFTPC . Pediatr Res 84, 419–425 (2018). https://doi.org/10.1038/pr.2018.30
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2018.30
This article is cited by
-
GCN5L1 regulates pulmonary surfactant production by modulating lamellar body biogenesis and trafficking in mouse alveolar epithelial cells
Cellular & Molecular Biology Letters (2023)
-
Surfactant protein disorders in childhood interstitial lung disease
European Journal of Pediatrics (2021)