Abstract
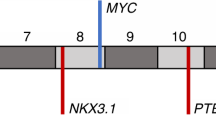
Four independent regions within 8q24 near the MYC gene are associated with risk for prostate cancer (Pca). Here, we investigated allelic imbalance (AI) at 8q24 risk variants and MYC gene DNA copy number (CN) in 27 primary Pcas. Heterozygotes were observed in 24 of 27 patients at one or more 8q24 markers and 27% of the loci exhibited AI in tumor DNA. The 8q24 risk alleles were preferentially favored in the tumors. Increased MYC gene CN was observed in 33% of tumors, and the co-existence of increased MYC gene CN with AI at risk loci was observed in 86% (P<0.004 exact binomial test) of the informative tumors. No AI was observed in tumors, which did not reveal increased MYC gene CN. Higher Gleason score was associated with tumors exhibiting AI (P=0.04) and also with increased MYC gene CN (P=0.02). Our results suggest that AI at 8q24 and increased MYC gene CN may both be related to high Gleason score in Pca. Our findings also suggest that these two somatic alterations may be due to the same preferential chromosomal duplication event during prostate tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA et al. A common variant associated with prostate cancer in European and African populations. Nat Genet 2006; 38: 652–658.
Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 2007; 39: 631–637.
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 2007; 39: 645–649.
Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet 2007; 39: 638–644.
Witte JS . Multiple prostate cancer risk variants on 8q24. Nat Genet 2007; 39: 579–580.
Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, Candreva A et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res 2007; 17: 1717–1722.
Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res 2007; 67: 2944–2950.
Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med 2008; 358: 910–919.
Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 2008; 100: 962–966.
Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007; 39: 984–988.
Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007; 39: 989–994.
Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 2008; 40: 1307–1312.
Kupfer SS, Torres JB, Hooker S, Anderson JR, Skol AD, Ellis NA et al. Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis 2009; 30: 1353–1357.
Tuupanen S, Niittymaki I, Nousiainen K, Vanharanta S, Mecklin JP, Nuorva K et al. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res 2008; 68: 14–17.
Qian J, Jenkins RB, Bostwick DG . Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Mod Pathol 1997; 10: 1113–1119.
Jenkins RB, Qian J, Lieber MM, Bostwick DG . Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997; 57: 524–531.
Liu W, Xie CC, Zhu Y, Li T, Sun J, Cheng Y et al. Homozygous deletions and recurrent amplifications implicate new genes involved in prostate cancer. Neoplasia 2008; 10: 897–907.
Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res 2007; 67: 8504–8510.
Ribeiro FR, Henrique R, Martins AT, Jeronimo C, Teixeira MR . Relative copy number gain of MYC in diagnostic needle biopsies is an independent prognostic factor for prostate cancer patients. Eur Urol 2007; 52: 116–125.
Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol 2008; 21: 1156–1167.
Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA et al. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 1996; 56: 3331–3337.
Liu W, Wu X, Zhang W, Montenegro RC, Fackenthal DL, Spitz JA et al. Relationship of EGFR mutations, expression, amplification, and polymorphisms to epidermal growth factor receptor inhibitors in the NCI60 cell lines. Clin Cancer Res 2007; 13: 6788–6795.
Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005; 6: 279–286.
Potts PR, Yu H . Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 2005; 25: 7021–7032.
Wirtenberger M, Hemminki K, Forsti A, Klaes R, Schmutzler RK, Grzybowska E et al. c-MYC Asn11Ser is associated with increased risk for familial breast cancer. Int J Cancer 2005; 117: 638–642.
Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 2009; 41: 885–890.
Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet 2009; 41: 455–459.
Acknowledgements
We thank all the men who volunteered to participate in this genetic study. This research was funded in part by the Department of Defense (DAMD W81XWH-07-1-0203 and DAMD W81XWH-06-1-0066).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chen, H., Liu, W., Roberts, W. et al. 8q24 allelic imbalance and MYC gene copy number in primary prostate cancer. Prostate Cancer Prostatic Dis 13, 238–243 (2010). https://doi.org/10.1038/pcan.2010.20
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pcan.2010.20
Keywords
This article is cited by
-
SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein
Oncogene (2017)
-
PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: implications for active surveillance
Modern Pathology (2016)
-
Analysis of genetic aberrations on chromosomal region 8q21–24 identifies E2F5 as an oncogene with copy number gain in prostate cancer
Medical Oncology (2013)