Abstract
Tumor cell heterogeneity poses a major hurdle in the treatment of cancer. Mammary cancer stem-like cells (MaCSCs), or tumor-initiating cells, are highly tumorigenic sub-populations that have the potential to self-renew and to differentiate. These cells are clinically important, as they display therapeutic resistance and may contribute to treatment failure and recurrence, but the signaling axes relevant to the tumorigenic phenotype are poorly defined. The zinc-finger transcription factor Kruppel-like factor 4 (KLF4) is a pluripotency mediator that is enriched in MaCSCs. KLF4 promotes RAS-extracellular signal-regulated kinase pathway activity and tumor cell survival in triple-negative breast cancer (TNBC) cells. In this study, we found that both KLF4 and a downstream effector, microRNA-206 (miR-206), are selectively enriched in the MaCSC fractions of cultured human TNBC cell lines, as well as in the aldehyde dehydrogenase-high MaCSC sub-population of cells derived from xenografted human mammary carcinomas. The suppression of endogenous KLF4 or miR-206 activities abrogated cell survival and in vivo tumor initiation, despite having only subtle effects on MaCSC abundance. Using a combinatorial approach that included in silico as well as loss- and gain-of-function in vitro assays, we identified miR-206-mediated repression of the pro-apoptotic molecules programmed cell death 4 (PDCD4) and connexin 43 (CX43/GJA1). Depletion of either of these two miR-206-regulated transcripts promoted resistance to anoikis, a prominent feature of CSCs, but did not consistently alter MaCSC abundance. Consistent with increased levels of miR-206 in MaCSCs, the expression of both PDCD4 and CX43 was suppressed in these cells relative to control cells. These results identify miR-206 as an effector of KLF4-mediated prosurvival signaling in MaCSCs through repression of PDCD4 and CX43. Consequently, our study suggests that a pluripotency factor exerts prosurvival signaling in MaCSCs, and that antagonism of KLF4-miR-206 signaling may selectively target the MaCSC niche in TNBC.
Similar content being viewed by others
Introduction
Compelling experimental evidence supports the hierarchical organization of certain human tumor types, including breast cancer.1, 2, 3, 4, 5, 6 These tumors comprise heterogenous mixtures of tumor cell populations that include cancer stem-like cells (CSCs), typically defined by their ability to initiate tumors in limiting dilution assays (that is, tumor-initiating cells, TICs). Furthermore, CSCs can survive and form tumorspheres in suspension culture, self-renew and differentiate.7, 8 These cells display resistance to chemotherapy, radiation therapy and other triggers of cell death, and are thought to contribute to cancer recurrence. Therefore, CSCs represent an important sub-population for therapeutic targeting.3, 5
In mammary carcinoma, functionally validated CSCs (termed MaCSCs (mammary CSCs)) have been identified by profiling the expression of cell surface markers such as PROCR (P) and ESA (E) and/or by assaying aldehyde dehydrogenase (ALDH) activity.9, 10, 11, 12 Despite this insight, the underlying mechanisms that mediate the MaCSC phenotype are unclear. For regulation of their abundance and/or their intrinsic properties such as resistance to cell death, several cellular signaling axes have been implicated including the WNT, NOTCH, TGFβ and SHH pathways.13, 14, 15
A potential mediator of the MaCSC phenotype is the pluripotency factor Kruppel-like factor 4 (KLF4). This zinc-finger transcription factor promotes the formation of induced pluripotent stem cells from adult somatic cells and can have both antitumorigenic and protumorigenic roles in a context-dependent manner.16, 17, 18, 19, 20 The capability of KLF4 to exert protumorigenic influences may reflect its role as a prosurvival stress response factor.21, 22, 23, 24, 25, 26, 27, 28 In support of a protumorigenic role, KLF4 promotes epithelial transformation in vitro, escape from RAS-induced senescence and skin tumor initiation in transgenic mice.16, 29, 30 Furthermore, loss-of-function studies reveal that KLF4 promotes cell survival following radiation-induced DNA damage, and promotes the tumorigenicity of colorectal CSCs-enriched spheroid cells.26, 31
In human breast cancer, KLF4 promoter demethylation and KLF4 protein expression indicate an unfavorable prognosis.32, 33, 34 KLF4 expression is positively correlated with tumor size, advanced grade and stage.35 We previously identified microRNAs, including microRNA-206 (miR-206) and miR-21, as direct transcriptional targets of KLF4 that promote RAS-extracellular signal-regulated kinase (ERK) signaling in triple-negative breast cancer (TNBC) cells.36, 37 Although on its own each miR exerts only subtle influences on RAS-ERK pathway activity, the coexpression of miR-206 and miR-21 potently represses the expression of pathway inhibitors including RASA1 and SPRED1. Furthermore, miR-206 directly represses KLF4 translation, constituting a feedback loop.36
In this study, we observed elevation of KLF4 and miR-206 in the P+/E+ and ALDHHigh MaCSC fractions. In TNBC cells, both KLF4 and miR-206 were critical for cell survival and in vivo tumor initiation. We identified the tumor-suppressor programmed cell death 4 (PDCD4) as a potential mediator of cell survival by miR-206. Furthermore, in TNBC cells we demonstrated the miR-206 regulation of a previously validated transcript, the gap junction protein connexin 43 (CX43/GJA1).38
Consistent with the elevated levels of miR-206 in MaCSCs, PDCD4 and CX43 levels were decreased. Supporting functional roles downstream of KLF4 and miR-206, suppression of either PDCD4 or CX43 led to anoikis resistance, an intrinsic property of CSCs.7, 39, 40, 41, 42, 43 Finally, further documenting a prosurvival role, miR-206 promoted chemoresistance of TNBC cells against paclitaxel or doxorubicin. Our studies identify KLF4 and miR-206 as functional MaCSC markers that mediate cell survival. Consequently, KLF4 and/or miR-206 may be therapeutically targeted to selectively cripple MaCSCs in TNBCs.
Results
miR-206 is highly expressed in basal-like breast cancers and MaCSCs
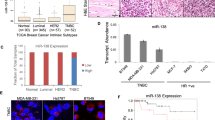
KLF4 protein levels correlate with an aggressive phenotype in breast tumors.32, 33, 35 Similar to KLF4, miR-206 was increased in human tumors of advanced histological grade (Figure 1a, left panel). Consistent with studies that identified upregulation of miR-206 in ER- breast tumors, miR-206 levels were elevated in TNBCs compared with both ER+ and HER2+ human subgroups (Figure 1a, right panel).44, 45 Enrichment of miR-206 was similarly observed in murine basal-like mammary tumors (Figure 1b). Compared with normal mammary tissues or tumors arising in the luminal MMTV_Neu model,46 we observed upregulation of both KLF4 and miR-206 in basal-like tumors derived from the C3(1)/SV40 large T antigen (C3(1)/TAg) genetically engineered mouse model (GEMM) (Figure 1c). These results are consistent with the direct regulation of miR-206 by KLF4 as previously reported.37
KLF4 and miR-206 are selectively expressed in basal-like mammary cancers and in the MaCSC population. (a) miR-206 levels were analyzed by microarray in 98 primary human breast tumors.91 The Gene Expression Omnibus (GEO) accession number is indicated. (Columns, mean; bars, s.e.m.; Hist. grade, histologic grade). (b) miR-206 levels were analyzed by microarray in 42 mammary tumors from GEMMs.90 The GEO accession number is indicated. (c) Klf4 and miR-206 expression was evaluated in normal mammary tissues from FVB/N mice and in primary tumors arising in the MMTV-Neu and C3(1)/TAg GEMMs. RNA levels were determined by qRT–PCR. (d) MaCSCs were isolated from MDA-MB-231 cells by sorting using PROCR (P) and ESA (E) as described.12 Transcript levels were analyzed in P+/E+ and P-/E- cells. (e) MaCSCs were isolated from SUM159PT cells and analyzed similarly as described above for MDA-MB-231 cells.53 For these cells, the P+ profile was used as a surrogate for CD44+/CD24-.10 (*P<0.05; **P<0.01; ***P<0.001).
MaCSCs are enriched in the triple-negative subgroup of breast cancer and are thought to contribute to the aggressive behavior of these cancers.47, 48, 49 Similar to human and murine mammary carcinoma cells displaying high ALDH activity,11, 50, 51, 52 MDA-MB-231 TNBC cells displaying the P+/E+ surface marker profile represent TICs.12 For SUM159PT cells, CD44+/CD24-/ESA+ subset was previously identified as TICs.53 As the P+ phenotype is a surrogate for the CD44+/CD24- profile, the P+/E+ SUM159PT cells are likely to represent MaCSCs.10
We analyzed KLF4 and miR-206 levels in flow-sorted sub-populations of MDA-MB-231 cells (Figure 1d, left panel). Compared with non-MaCSCs (that is, P-/E-), miR-206 and KLF4 were increased in the P+/E+ sub-population (Figure 1d, middle panels). Using P+/E+ cells, we profiled the expression of other genes associated with stem-like cell phenotypes.9, 14, 18, 19 Compared with P-/E- cells, the expression of CD44, MYC, SOX2, NANOG, ZEB1 and SNAI2 was upregulated in P+/E+ cells, whereas CD24 and POU5F1 (OCT3/4) expression were decreased (Figure 1d, right panel). Similarly, the P+/E+ fraction of SUM159PT cells displayed elevated levels of KLF4 and miR-206, and showed a similar stem cell marker profile as the MDA-MB-231 cells (Figure 1e). These results associate KLF4 and miR-206 with the MaCSC phenotype in human breast cancer models.
KLF4 and miR-206 are enriched in MaCSCs derived from human patient-derived xenografts (PDXs) and the C3(1)/TAg GEMM
KLF4 was similarly consistently elevated in lineage-negative (Lin-)/ALDHHigh MaCSCs isolated from human mammary tumor tissues that were passaged as PDXs (Figure 2a). miR-206 was upregulated in three of these four cases. Notably, none of these tumors displayed an appreciable CD44+/CD24- MaCSC population (data not shown), consistent with the variable expression of these markers in patient samples.10, 54, 55
KLF4 and miR-206 are enriched in ALDHHigh MaCSCs derived from human PDXs and the C3(1)/TAg GEMM. (a) KLF4 and miR-206 levels were measured in MaCSCs purified in replicate manner from four cases of human mammary carcinoma passaged as xenografts in mice (PDXs). Purified lineage-negative (Lin-) cells were sorted based on ALDH activity. Fluorescence was analyzed in the presence of the ALDH substrate BAAA and in presence/absence of the ALDH inhibitor DEAB. The number of xenografted tumors that were analyzed for each case is indicated below the column data (bars, s.e.m.). n.s., not significant. (b) Lin- cells were isolated from the spontaneous mammary tumors arising in C3(1)/TAg females (N=3). Cells from each animal were grown as either adherent monolayers (2D) or in suspension (3D) for 7–10 days. Photomicrographs (left) depict the morphology of cultured cells. RNA was extracted from 2D or 3D cell cultures and Klf4 and miR-206 levels were determined. (c) Klf4 and miR-206 levels were measured in the ALDHHigh tumor cells isolated from C3(1)/TAg animals (N=3, see panel b). ALDHHigh tumor cells derived from other GEMMs of mammary cancer have been demonstrated to be enriched for TICs50, 52 (*P<0.05; **P<0.01; ***P<0.001).
Tumorspheres are enriched for MaCSCs.7, 8 Compared with cells grown in adherent (two-dimensional (2D)) monolayers, tumorspheres formed from the Lin- cells of C3(1)/TAg mammary tumors showed elevated levels of Klf4 and miR-206 (Figure 2b). ALDHHigh cells from other mammary cancer GEMMs were previously shown to have properties of MaCSCs.50, 52 Similar to the human tumors, Lin-/ALDHHigh cells of C3(1)/TAg mammary tumors also had increased Klf4 and miR-206 relative to ALDHLow cells (Figure 2c). These results identify KLF4 and miR-206 as MaCSC markers and potential mediators of MaCSC malignant properties.
KLF4 and miR-206 can promote MaCSC abundance
To determine the effect of KLF4-miR-206 signaling on MaCSC abundance, we depleted KLF4 in MDA-MB-231 cells using two distinct lentiviral short hairpin RNA constructs (Figure 3a, left upper panel). Consistent with previous studies, miR-206 was suppressed following KLF4 knockdown (Figure 3a, left lower panel). In addition, P+/E+ cell abundance was modestly decreased upon KLF4 depletion (Figure 3a, middle and right panels). Conversely, gain-of-function experiments showed that exogenous KLF4 promoted both miR-206 levels and the abundance of P+/E+ cells (Figure 3b).
KLF4 and miR-206 promote MaCSC abundance. (a) MDA-MB-231 cells were transduced with lentiviral vectors expressing KLF4 short hairpin RNAs or a non-targeting control (Ctl). KLF4 protein expression was analyzed by immunoblot (left upper panel). β-Actin served as a loading control. miR-206 levels were measured by stem loop qRT–PCR (left lower panel). The cell surface marker profile of the transduced cells was analyzed by flow cytometry (representative scatter plot, middle panel; column data, right panel) (N=3; bars, s.e.m.). (b) MDA-MB-231 cells were transduced with a retroviral vector encoding KLF4 or empty vector (Ctl). KLF4 and miR-206 levels were analyzed in these cells (left panels) and the MaCSC abundance was determined by flow cytometry. (c) MDA-MB-231 cells were transfected with either miR-206 mimic or control oligonucleotides (Ctl) and then analyzed as in the previous panels. (d) SUM159PT cells were transfected with the indicated miR-mimics and then analyzed as in the previous panels. (e) MDA-MB-231 cells were transfected with miR-206 mimic or control. The indicated number of cells were mixed with matrigel (50% (vol/vol) in DMEM) and injected into NSG mice. Tumor initiation was measured at 4 weeks post-injection (**P<0.01; ***P<0.001).
We next sought to determine whether miR-206 could have a causal role downstream of KLF4 to regulate MaCSC abundance. As expected, transfection of miR-206 mimic into MDA-MB-231 cells elevated the miR-206 level as detected by quantitative reverse transcription and PCR (qRT–PCR; Figure 3c, left upper panel). In addition, the level of KLF4 was suppressed, attributed to direct regulation of KLF4 protein translation by miR-206 (Figure 3c, left lower panel).36 Despite the reduced levels of KLF4, miR-206-transfected cells displayed higher P+/E+ cell abundance relative to the control cells (Figure 3c, right panel). Similar regulation of P+/E+ cell abundance by miR-206 was observed for SUM159PT cells (Figure 3d). These results establish miR-206 as a potential effector of KLF4 for regulation of MaCSC abundance.
To determine whether miR-206 can promote the MaCSC phenotype, we assayed by limiting dilution the capability of miR-206-transfected MDA-MB-231 cells to initiate tumors in vivo. Consistent with an increased number of P+/E+ cells, miR-206-transfected cells formed tumors more efficiently in NOD/SCID-gamma (NSG) mice compared with control cells (Figure 3e; 2 × 103 cells, P=0.0022). These results implicate miR-206 as an effector of KLF4 that promotes tumor initiation.
Endogenous KLF4 and miR-206 promote tumor cell survival and in vivo tumorigenesis
We next examined the impact of endogenous KLF4-miR-206 signaling on tumor initiation. Depletion of KLF4 reduced the tumor initiation rate of MDA-MB-231 cells in athymic nude mice (Figure 4a, left panels). This decrease in tumor incidence was reflected by the reduced mean tumor volume for all animals combined (Figure 4a, middle panels). Indicating that the major effect of KLF4 in this setting is restricted to tumor initiation, analysis of the tumor-positive subset revealed little difference in the tumor growth rate between KLF4-depleted cells and the control (Figure 4a, right panels).
Endogenous KLF4-miR-206 signaling promotes in vivo tumorigenesis and cell survival. (a) KLF4-depleted and control MDA-MB-231 cells were orthotopically injected into athymic nude mice. Tumor initiation and tumor size were determined twice per week using digital calipers (right panels; bars, s.e.m.). (b) MDA-MB-231 cells were transfected with the indicated anti-miRs. Briefly, cells were subjected to sequential transfections in vitro. At 2 days post-transfection, the cells were injected into athymic nude mice. Tumor incidence and growth were measured as described above. (c) Residual transfected cells (see panel b) were directly lysed for immunobot analysis (left panel) or else placed in culture for 2D cell proliferation analysis (right panel, ATPlite; N=6, bars, s.d.). Post-tf, post-transfection. (d) TNBC cells were transfected with either anti-miR-206 or anti-miR-Ctl and then deprived of matrix for 24 h. Anoikis was measured by Trypan blue exclusion (left panel, N=3, bars, s.d.). In parallel, cells were assayed by immunoblot analysis of cleaved PARP. Cyclohexamide (CHX) treatment served as a positive control for induction of cell death. (e) Anoikis was measured in KLF4-depleted MDA-MB-231 cells or control cells by Trypan blue exclusion (N=3, bars, s.d.), by flow cytometric analysis of propidium iodide (PI)-stained cells (N=3, bars, s.d.), and by analysis of cleaved PARP. n.s., not significant (*P<0.05; **P<0.01; ***P<0.001).
To study the role of endogenous miR-206 during in vivo tumorigenesis, we analyzed the tumorigenicity of MDA-MB-231 cells treated by in vitro transfection of anti-sense oligonucleotides (anti-miR-206). Compared with cells transfected with the control, anti-miR-206 treatment reduced both tumor incidence and tumor growth (Figure 4b). As an indicator of successful transfection, KLF4 expression was increased (Figure 4c, left panel). Effects on tumor growth were not likely attributed to differences in cell proliferation rates, as anti-miR-206 had little effect (Figure 4c, right panel).
The critical role of endogenous miR-206 for tumor initiation following orthotopic injection, despite its minimal effects on cell proliferation or MaCSC abundance, pointed to a potential role in regulating cell survival. We therefore assayed for resistance to cell death following matrix deprivation (anoikis), an intrinsic property of CSCs.7, 39, 40, 41, 42, 43 Indeed, consistent with our previous report that analyzed two human TNBC cell lines,37 anti-miR-206 transfection sensitized several human TNBC models and a murine basal-like mammary cancer model (that is, M6 cells) to anoikis (Figure 4d, left panel). Consistent results were obtained when anoikis was analyzed by poly ADP ribose polymerase (PARP) cleavage (Figure 4d, right panel). In support of a prosurvival role for endogenous miR-206, depletion of KLF4 sensitized TNBC cells to anoikis (Figure 4e). These results suggest that endogenous KLF4 exerts a prosurvival effect by induction of miR-206.
miR-206 suppresses the translation of the tumor-suppressor PDCD4 and promotes tumor cell survival
We previously reported that RAS-ERK signaling, a prosurvival pathway, is maintained in TNBC cells by KLF4, in part through its regulation of miR-206.37 In contrast to the prominent effect of miR-206 on tumor initiation and cell survival, on its own this miR has only limited effects on ERK activity.37 We therefore sought to better understand how endogenous miR-206 can promote anoikis resistance.
The tumor-suppressor PDCD4 was identified as a potential miR-206 targeted transcript by multiple miR-target prediction tools.37, 56 PDCD4 is a negative regulator of RAS-ERK-AP1 signaling and protein translation, and promotes breast cancer cell apoptosis.57, 58, 59 We therefore analyzed PDCD4 as a miR-206-regulated transcript.
Consistent with regulation of PDCD4 by miR-206, KLF4 depletion in MDA-MB-231 cells increased PDCD4 expression (Figure 5a, left panel). Similarly, although anti-miR-206 treatment elevated PDCD4, transfection of miR-206 mimic was suppressive (Figure 5a, middle and right panels). Direct regulation of PDCD4 by miR-206 was determined using translational reporter assays. Fragments of the PDCD4 3′ UTR containing two putative miR-206-binding sites (denoted WT-A and WT-B; Figure 5b) were cloned downstream of the open reading frame of firefly luciferase (luc). Relative to the controls, in MDA-MB-231 cells miR-206 mimic repressed WT-reporter luc activity by 72%, and anti-miR-206 induced the reporter by 1.9-fold (Figure 5c). Reporter regulation by miR-206 was abolished by mutation of site WT-A, but not by mutation of site WT-B, thus identifying site WT-A as a functional miR-206-binding site (Figure 5b and c). In agreement with previous studies, PDCD4 depletion in TNBC cells promoted resistance to anoikis, with little or no effect on 2D proliferation (Figure 5d).
miR-206 suppresses the translation of the tumor-suppressor PDCD4. (a) PDCD4 levels were determined by immunoblot analysis of the indicated cells. (b) Alignment of the PDCD4 3′UTR region indicating two potential miR-206 binding sites, WT-A and WT-B. The miR-206 seed sequence is underlined. Mutated miR-206 binding sites in the PDCD4 3′ UTR that were utilized in translational reporter assays are indicated (mt206-A and mt206-B). (c) For analysis of PDCD4 protein translation, MDA-MB-231 cells were co-transfected with reporters in combination with either miR-mimic (left panel) or anti-miR (right panel). The normalized activity of the reporters relative to empty luc vector was analyzed 24 h post-transfection (N=3; bars, s.e.m.). (d) PDCD4 was depleted in the indicated TNBC cells and PDCD4 levels were determined by immunoblot (upper panels). Cells were suspended in 3D culture for 24 h, and anoikis was measured by flow cytometric analysis of propidium iodide (PI)-stained cells (middle panels; N=3; bars, s.e.m.). Following 4 days of 2D culture, the relative cell number of PDCD4-depleted cells and control cells was determined by the ATPlite assay (N=6; bars, s.d.). (e) PDCD4 mRNA and protein expression was analyzed in the indicated sub-populations of TNBC cells. Non-MaCSCs comprises the P+/E- and P-/E- subgroups. The immunoblot data correspond to one of the three independent experiments that analyzed mRNA levels (N=3, bars, s.e.m.). (f) MaCSC abundance was analyzed in PDCD4-depleted TNBC cells and control cells (N=3; bars, s.d.). n.s., not significant (*P<0.05; **P<0.01; ***P<0.001).
Consistent with miR-206 regulation of PDCD4 in MaCSCs, the P+/E+ sub-population of MDA-MB-231 cells exhibited decreased levels of PDCD4 mRNA and protein compared with non-MaCSCs (Figure 5e). In TNBC cells, the depletion of PDCD4 was not sufficient to alter the abundance of the P+/E+ fraction (Figure 5f). These results appear to support a selective role of PDCD4 for suppression of tumor cell survival.
miR-206 promotes cell survival by suppressing CX43 in MaCSCs
Our identification of miR-206 regulation of PDCD4 led us to seek additional targets of this miR that may be important for promoting cell survival. DIANA-miRPath analysis identifies gap junction signaling as the top-ranked miR-206-regulated pathway (P=2.58 × 10−6).60 Among the targeted gap junction proteins, CX43 is a validated miR-206-regulated transcript, as previously shown in muscle cells.38, 61 CX43 is deficient in human breast tumor cells and MaCSCs, and may exert a tumor-suppressor role.62, 63, 64, 65, 66, 67, 68
Consistent with its regulation by miR-206 in breast cancer cells, CX43 was increased in KLF4-depleted MDA-MB-231 cells (Figure 6a, left panel). Similarly, inhibition of miR-206 led to elevated CX43 levels, and transfection of miR-206 mimic was suppressive (Figure 6a, middle and right panels). In TNBC cells, the activity of a translational reporter containing the CX43 3’ UTR was induced by 1.5-fold following anti-miR-206 treatment, and suppressed by 53% following transfection of miR-206 mimic (Figures 6b and c). Supporting the direct regulation of CX43 by miR-206 in breast tumor cells, mutation of site A (mut206-A) abolished regulation by miR-206 (Figure 6c). Similar to PDCD4 depletion, suppression of CX43 in TNBC cells promoted resistance to anoikis, with only subtle effects on cell proliferation (Figure 6d).
KLF4-miR-206 signaling suppresses CX43 in MaCSCs. (a) CX43 expression was analyzed in shKLF4 cells and control cells by immunoblot. Similarly, CX43 expression was analyzed in cells transfected with the indicated miR mimic or anti-miR. (b) Alignment of CX43 3’UTR region, indicating two previously validated miR-206 binding sites, WT-A and WT-B.38 The miR-206 seed sequence is underlined. The mutation generated in miR-206 binding site A is indicated (mt206-A). (c) For analysis of CX43 protein translation, MDA-MB-231 cells were co-transfected with reporters in combination with anti-miR (left panel) or miR-mimic (right panel). The normalized activity of the reporters relative to empty luc vector was analyzed at 24 h post-transfection. (d) CX43 expression was assayed in CX43-depleted TNBC cells and control cells (upper panels). Cells were suspended in 3D culture for 24 h, and anoikis was measured by flow cytometric analysis of propidium iodide (PI)-stained cells (middle panels, N=3; bars, s.e.m.). Following 4 days of 2D culture, the relative cell number of CX43-depleted cells and control cells was determined by the ATPlite assay (lower panels, N=6; bars, s.d.). (e) CX43 mRNA and protein expression was analyzed in the indicated sub-populations of MDA-MB-231 cells (N=3; bars, s.e.m.). Non-MaCSCs were composed of the P+/E- and P-/E- subgroups. n.s., not significant (*P<0.05; **P<0.01; ***P<0.001).
Compared with the non-MaCSC fraction, P+/E+ MDA-MB-231 cells displayed lower CX43 mRNA and protein (Figure 6e). These results support a previous study that reported low CX43 expression in mammary TICs.64 Similarly to PDCD4, knockdown of CX43 did not consistently alter the P+/E+ cell abundance in TNBC cells, suggesting a selective role in tumor cell survival (data not shown).
miR-206 confers chemoresistance in TNBC cells
Consistent with the promotion of cell survival by miR-206 as determined by anoikis assays, TNBC cells transfected with miR-206 mimic were more resistant to paclitaxel or doxorubicin (Figure 7a). Furthermore, inhibition of the endogenous miR-206 moderately sensitized TNBC cells to either agent (Figure 7b). Collectively, these results link pluripotency factor signaling and the enhanced cell survival of MaCSCs, supporting roles of KLF4-miR-206 signaling for breast tumor cell survival, chemoresistance, and tumor initiation through the repression of PDCD4 and CX43 (Figure 7c).
miR-206 promotes chemoresistance in TNBC cells. (a, b) TNBC cells were transfected as indicated. At 48 h post-transfection, cells were treated with the indicated concentrations of paclitaxel or doxorubicin for a duration of 72 h. Cell viability was determined by the ATPlite assay (N=6; bars, s.d.). (c) Schematic of KLF4-miR-206 regulation of PDCD4 and CX43. (**P<0.01; ***P<0.001).
Discussion
CSCs were first identified in hematopoietic malignancies and subsequently in solid tumors such as breast cancer.1, 3, 5, 69 Despite substantial progress, questions remain regarding the relationship of CSCs to the adult stem cells of normal tissue, and the nature of the signaling pathways that regulate CSC properties.6 Despite this uncertainty, it is clear that CSCs represent a highly malignant sub-population of tumor cells with the capability to resist therapy.3, 5
In TNBC cells, KLF4 directly regulates miR-206 transcription, and depletion of KLF4 consistently results in loss of the vast majority of miR-206.36, 37 In this study, we identified KLF4 and miR-206 as critical promoters of breast tumor cell survival. Both factors were preferentially expressed in the MaCSCs purified from 2D cell culture models of TNBC, from tumorspheres cultured in 3D, from human PDXs and from primary mouse mammary cancers. As shown by anti-miR treatment of TNBC cells, endogenous miR-206 directly repressed the translation of the tumor suppressors PDCD4 and CX43 and promoted tumor cell survival, chemoresistance and in vivo tumor initiation. Immunoblot analysis of MaCSCs revealed suppressed levels of both PDCD4 and CX43. Mirroring the role of endogenous miR-206, depletion of each tumor suppressor did not alter the abundance of CSCs, but instead enhanced tumor cell survival consistent with previous reports.57, 70
miRs can act as critical factors for regulating the abundance and/or survival of MaCSCs.71, 72, 73, 74 In skeletal muscle, miR-206 is important for repression of PAX7 during stem cell differentiation, and for muscle regeneration following injury.75, 76, 77, 78, 79 In a mammary cancer context, miR-206 expression is elevated in ER- tumors, which are enriched for MaCSCs.44, 45, 47, 48, 49 In agreement with previous studies, we observed that miR-206 is upregulated in human breast cancers that display a higher grade, in human TNBCs and in basal-like mammary tumors derived from the C3(1)/TAg GEMM (Figure 1a and c).
Multiple previous studies have reported that enforced expression of miR-206 can suppress tumor cell proliferation, invasion or metastasis.45, 80, 81, 82, 83 These tumor-suppressor-like effects of miR-206 may result from higher level enforced expression of the exogenous miR. In this study, suppression of endogenous miR-206 blocked tumor initiation, and moderate (fivefold) overexpression of exogenous miR-206 promoted initiation in a limiting dilution assay. In addition, we observed that either exogenous or endogenous miR-206 could promote malignant properties including tumor cell survival and drug resistance.
Depletion of endogenous KLF4 suppressed in vivo tumor initiation by MDA-MB-231 cells in athymic nude mice, yet had little effect on the growth rate of established tumors. Similarly as observed for KLF4, transient inhibition of endogenous miR-206 by anti-miR-206 transfection suppressed tumor initiation in vivo but did not alter the in vitro proliferation or the MaCSC abundance. These results suggest that endogenous KLF4 can signal through miR-206 to promote tumor initiation, probably by impacting cell survival rather than MaCSC abundance. In contrast, exogenous KLF4 or miR-206 promoted MaCSC abundance, mirroring the role of exogenous KLF4 for generation of induced pluripotent stem cells.18, 19 It will be interesting to determine whether miR-206 similarly influences the generation of induced pluripotent stem cells.
In this study, we have identified endogenous KLF4 and a downstream effector, miR-206, as functional markers and prosurvival factors that are enriched in MaCSCs. Prosurvival signaling by miR-206 was attributed to direct regulation of PDCD4 and CX43, and miR-206 enhanced the chemoresistance of TNBC cells. Our study, therefore, provides a rationale for miR-206-directed antago-miR therapy for the sensitization of the MaCSCs.74, 84, 85, 86, 87, 88
Materials and methods
Cell lines, cell culture and drug treatments
MDA-MB-231 cells were provided by Katri S Selander (University of Alabama at Birmingham, AL, USA), SUM159PT cells were provided by Gary L Johnson (University of North Carolina at Chapel Hill, NC, USA) and M6 mammary carcinoma cells derived from the C3(1)/SV40 TAg mouse model were provided by Jeffrey E Green (NIH). HCC1143 cells were from ATCC (Manassas, VA, USA). Cells were maintained as subconfluent monolayers as previously described.36, 37
For chemoresistance experiments, cells were treated with the indicated doses of paclitaxel (Sigma, St Louis, MO, USA) or doxorubicin (Merck, Billerica, MA, USA) for 72 h. Cells were treated with cycloheximide (Sigma) at 20 μg/ml for 24 h. Cell proliferation was determined using the ATPlite Luminescence Assay System (PerkinElmer, Waltham, MA, USA).
Retroviral transduction
Suppression studies utilized the following pGIPZ lentiviral shRNAmir plasmids (V2LHS_28277 – shKLF4-1, V3LHS_410934 – shKLF4-2, V3LHS_411731 – shCX43-1, V3LHS_411733 – shCX43-2, V3LHS_366084 – shPDCD4-1, V3LHS_366087 – shPDCD4-2; GE Dharmacon/Open Biosystems, Lafayette, CO, USA). The retroviral vector pBABEpuro-HA-KLF4 and viral transduction was previously described.36 Cells were selected using puromycin (1 μg/ml).
Plasmid construction
pMIR-REPORT firefly luciferase vector was purchased from Ambion (Austin, TX, USA). pRL-TK Renilla luc reporter was obtained from Promega (Madison, WI, USA).
Complementary DNA clones containing fragments of the 3′ UTR of PDCD4 (clone ID: NM_014456) and CX43/GJA1 were purchased from Open Biosystems and OriGene (Rockville, MD, USA), respectively.
To construct a WT PDCD4 translational reporter, a 1.7-kb fragment representing the 3′ UTR was excised using MluI and inserted into MluI-digested pMIR-REPORT. To construct a WT CX43/GJA1 translational reporter, a 1.7-kb fragment representing the CX43 3′ UTR was generated by sequential treatment with EcoRI, Klenow fragment and MluI. This fragment was inserted into pMIR-REPORT vector that was prepared by sequential treatment with SacI, Klenow fragment and MluI.
PDCD4 and CX43 reporters with mutation in the miR-seed complementary regions were generated by PCR mutagenesis. Oligonucleotides are listed in Supplementary Table S1. WT reporters were mutated so as to conserve the predicted secondary structure of the 3′ UTR.89 Cloned PCR products were confirmed by sequence analysis.
Transient transfection and translation reporter assays
The following anti-miR inhibitors (AM) and miR-mimics (PM) were obtained from Ambion and diluted to 20 μM in nuclease-free water: hsa-miR-206 (AM10409, PM10409), AM-negative control (AM17010), and PM-negative control (AM17110). Cells were subjected to reverse transfection and, 24 h later, forward transfection was performed as described.36 At 24 h after the start of the forward transfection, cell extracts were prepared for expression studies, or cells were used for phenotypic studies. Translational reporter assays were performed following just one transfection, at 24 h after the start of the reverse transfection. Inhibitors/mimics were co-transfected with reporter plasmids, and Dual-Luciferase Reporter Assays (Promega) were performed as described.36
Immunoblot analysis and antibodies
Cell extracts for immunoblot analysis were prepared as previously described.36 PARP cleavage assays were performed as recommended (Roche, Indianapolis, IN, USA). Following electrophoresis, proteins were transferred onto nitrocellulose membranes and probed with the indicated antibody: KLF4 (Santa Cruz Biotechnology, Dallas, TX, USA), PDCD4 (Rockland Immunochemicals, Philadelphia, PA, USA), CX43 (Sigma), PARP (Roche) or β-actin (Santa Cruz Biotechnology). Bound antibodies were detected using Pierce ECL Western Blotting Substrate (ThermoFisher Scientific, Waltham, MA).
Animal studies
Female athymic nude mice (Crl:NU(NCr)-Foxn1nu, Charles River, Frederick, MD, USA) were obtained at 6–8 weeks of age. In all, 2 × 106 cells were suspended in Dulbecco’s modified Eagle’s medium (DMEM) and injected into the fourth mammary fat pad. For tumor initiation/limiting dilution assays, NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson Lab, Bar Harbor, ME, USA) were obtained at 6–8 weeks of age. Tumor cells were suspended in DMEM containing matrigel (50% (vol/vol)) and injected into the fourth mammary fat pad. Tumors were measured semiweekly using digital calipers. Tumor volume was determined by π(L1 × L22)/6 (L1, long axis; L2, short axis), and tumor initiation was defined as ⩾2 mm for both L1 and L2. Animal procedures were performed under an approved protocol.
Isolation of mammary carcinoma cells from tumors
Human mammary cancer tissue was passaged as PDXs in NSG mice. HCI-001 and HCI-002 were obtained from Alana L Welm, University of Utah, and PEN-025 and PEN-027 were obtained from the West Virginia University Tissue Bank. PDX tumors and tumors arising in female C3(1)/TAg mice were harvested upon reaching a size of 1-2 cm3. To isolate mammary carcinoma cells, tumors were minced and suspended in DMEM/F12 containing Gentle Collagenase/Hyaluronidase (STEMCELL Technologies, Vancouver, BC, Canada) and then processed as recommended by the manufacturer. Briefly, tumor cell suspensions were incubated with mild agitation at 37 °C for 15 h. Red blood cells were lysed by the addition of 0.16 M Tris-NH4Cl (pH 7.6) and incubation at 25 °C for 3 min. Red blood cell lysis was stopped by the addition of DMEM/F12 containing 10% fetal bovine serum . The suspension was centrifuged and the resulting cell pellet was washed twice with DMEM/F12 containing 10% fetal bovine serum , resuspended in Trypsin-EDTA (0.25%; Media Tech, Corning, NY, USA) for 3 min with disaggregation by pipette, and then washed once again. Cells were resuspended in dispase and DNAse I (STEMCELL Technologies) at final concentrations of 4.2 mg/ml and 192 μg/ml, respectively. The cells were centrifuged and the cell pellet was resuspended in Hank’s balanced salt solution containing 10 mM HEPES-KOH, pH 7.2 and 2% fetal bovine serum .
Depletion of lineage-positive (Lin+) cells from prepared tumor cell suspensions was performed using an AutoMACS sorter (Miltenyi Biotec, San Diego, CA, USA). Briefly, cells were suspended in ice cold staining buffer (phosphate-buffered saline supplemented with 0.5% (wt/vol) bovine serum albumin) and blocked with 10 μg/ml mouse immunoglobulin G (Sigma) for 15 min. Cells were stained with the following biotin-conjugated antibodies (BD Bioscience, San Jose, CA, USA): anti-mouse-CD31 (clone 390), anti-CD45 (clone 30-F11), anti-TER-119 (clone TER-119). Anti-CD140b was from eBioscience, San Diego, CA, USA (clone APB5). Cells were washed with labeling buffer (phosphate-buffered saline, pH 7.2 containing 0.5% bovine serum albumin and 2 mM EDTA) and incubated with streptavidin microbeads (Miltenyi Biotec) before magnetic cell sorting as recommend by manufacturer.
Analysis and purification of MaCSCs
For analysis of PROCR/ESA expression, cells were blocked with 10 μg/ml normal human immunoglobulin G (R&D System, Minneapolis, MN, USA) in ice cold staining buffer (phosphate-buffered saline supplemented with 1% (vol/vol) fetal bovine serum) for 15 min. Cells were stained with anti-human PROCR-APC (clone RCR-227; eBioscience) and anti-human ESA-PerCP-Cy5.5 (clone EBA-1; BD Bioscience). Cells were centrifuged at 300 × g for 5 min at 4 °C, and washed twice with staining buffer before analysis.
ALDH activity was evaluated by flow cytometry using the ALDEFLUOR assay (STEMCELL Technologies). Cell sorting or flow cytometry was performed on a BD FACSAria using BDFACSDiva software version 6.1, or on a BD Fortessa using BDFACSDiva software version 7.0 (Becton Dickinson, San Jose, CA, USA). For analysis, a minimum of 10 000 events were collected for each sample. The data were analyzed by using FCS Express 4 Research Edition software (De Novo software, Glendale, CA, USA).
Tumorsphere formation and anoikis assays
To grow tumorspheres, 2 × 104 Lin- cells were placed in suspension cultures in low attachment plates (Costar, Corning, NY, USA) using DMEM/F12 supplemented with B27, 4 μg/ml heparin, 20 ng/ml epidermal growth factor, 20 ng/ml fibroblast growth factor and 1% (wt/vol) methylcellulose. For analysis of anoikis, cells were suspended in culture as previously described.37 Cell death was analyzed by propidium iodide staining and flow cytometry (Invitrogen, Carlsbad, CA, USA), by Trypan blue exclusion, or by analysis of cleaved PARP.
Expression analyses
Microarray data were extracted from GEO accessions GSE45666 and GSE23978 and then normalized to the geometric median.90, 91
For qRT–PCR, total RNA was extracted and mRNA and miR levels were analyzed as previously described.36 Reactions were normalized to B2M or RPLP0 for mRNA analysis, or to U6 snRNA for miR analysis. Primer sequences are listed in Supplementary Table S2. PCR reactions were performed on a Mx3005P Real-Time PCR System (Stratagene, La Jolla, CA, USA). mRNA and miR levels were determined by the ΔΔCT method.92 For all RNA measurements, three independent experiments were performed in duplicate manner.
Statistical analysis
Data were analyzed using either the unpaired t-test (two-tailed), or else one-way analysis of variance followed by Tukey’s multiple comparison ad hoc post-test. Tumor volumes were analyzed using two-way analysis of variance with a Bonferroni post-test. Tumor initiation was analyzed using a 2 × 2 contingency table with a Fisher’s exact test. Statistical analyses were performed in GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA. Differences were considered significant when the analysis yielded P<0.05.
References
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Hwang-Verslues WW, Chang KJ, Lee EY, Lee WH . Breast cancer stem cells and tumor suppressor genes. J Formos Med Assoc 2008; 107: 751–766.
Visvader JE, Lindeman GJ . Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008; 8: 755–768.
Shackleton M, Quintana E, Fearon ER, Morrison SJ . Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 2009; 138: 822–829.
Clevers H . The cancer stem cell: premises, promises and challenges. Nat Med 2011; 17: 313–319.
Beck B, Blanpain C . Unravelling cancer stem cell potential. Nat Rev Cancer 2013; 13: 727–738.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003; 17: 1253–1270.
Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS . Stem cells in normal breast development and breast cancer. Cell Prolif 2003; 36 (Suppl 1): 59–72.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003; 100: 3983–3988.
Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J et al. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007; 11: 259–273.
Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1: 555–567.
Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH, Tsai ST et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS ONE 2009; 4: e8377.
Reya T, Clevers H . Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–850.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Karamboulas C, Ailles L . Developmental signaling pathways in cancer stem cells of solid tumors. Biochim Biophys Acta 2013; 1830: 2481–2495.
Rowland BD, Bernards R, Peeper DS . The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005; 7: 1074–1082.
Rowland BD, Peeper DS . KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer 2006; 6: 11–23.
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872.
McConnell BB, Yang VW . Mammalian kruppel-like factors in health and diseases. Physiol Rev 2010; 90: 1337–1381.
McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci USA 2001; 98: 8955–8960.
Pedersen TX, Leethanakul C, Patel V, Mitola D, Lund LR, Dano K et al. Laser capture microdissection-based in vivo genomic profiling of wound keratinocytes identifies similarities and differences to squamous cell carcinoma. Oncogene 2003; 22: 3964–3976.
Yoon HS, Chen X, Yang VW . Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem 2003; 278: 2101–2105.
Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK . Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem 2005; 280: 9719–9727.
Liu Y, Wang J, Yi Y, Zhang H, Liu J, Liu M et al. Induction of KLF4 in response to heat stress. Cell Stress Chaperones 2006; 11: 379–389.
Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW . Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene 2007; 26: 2365–2373.
Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J et al. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem 2007; 282: 13769–13779.
Liao X, Haldar SM, Lu Y, Jeyaraj D, Paruchuri K, Nahori M et al. Kruppel-like factor 4 regulates pressure-induced cardiac hypertrophy. J Mol Cell Cardiol 2010; 49: 334–338.
Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, Kie-Bell P, Grizzle W et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ 1999; 10: 423–434.
Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 2005; 24: 1491–1500.
Leng Z, Tao K, Xia Q, Tan J, Yue Z, Chen J et al. Krüppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS ONE 2013; 8: e56082.
Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res 2004; 10: 2709–2719.
Chu PY, Hsu NC, Liao AT, Yeh KT, Hou MF, Liu CH et al. Elevated Kruppel-like factor 4 transcription factor in canine mammary carcinoma. BMC Vet Res 2011; 7: 58.
Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A et al. DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol 2011; 5: 77–92.
Chen CJ, Lin SE, Lin YM, Lin SH, Chen DR, Chen CL et al. Association of expression of kruppel-like factor 4 and kruppel-like factor 5 with the clinical manifestations of breast cancer. Pathol Oncol Res 2012; 18: 161–168.
Lin CC, Liu LZ, Addison JB, Ivanov AV, Ruppert JM . A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol 2011; 31: 2513–2527.
Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM et al. microRNAs-206 and -21 cooperate to promote RAS-ERK signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol 2014; 34: 4143–4164.
Anderson C, Catoe H, Werner R . MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 2006; 34: 5863–5871.
Reynolds BA, Weiss S . Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol 1996; 175: 1–13.
Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA 2000; 97: 14720–14725.
Dontu G, Wicha MS . Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia 2005; 10: 75–86.
Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res 2010; 70: 709–718.
Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA 2011; 108: 7950–7955.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–7070.
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H . miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res 2008; 68: 5004–5008.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8: R76.
Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res 2008; 10: R53.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 2009; 106: 13820–13825.
Perou CM . Molecular stratification of triple-negative breast cancers. Oncologist 2010; 15 (Suppl 5): 39–48.
Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ . A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev 2007; 21: 3238–3243.
Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 2009; 69: 1302–1313.
Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S et al. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res 2009; 69: 466–474.
Fillmore CM, Kuperwasser C . Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 2008; 10: R25.
Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K . Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res 2010; 16: 876–887.
Giatromanolaki A, Sivridis E, Fiska A, Koukourakis MI . The CD44+/CD24- phenotype relates to 'triple-negative' state and unfavorable prognosis in breast cancer patients. Med Oncol 2011; 28: 745–752.
Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS ONE 2012; 7: e42390.
Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH . Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 2004; 23: 8135–8145.
Lankat-Buttgereit B, Goke R . The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell 2009; 101: 309–317.
Santhanam AN, Baker AR, Hegamyer G, Kirschmann DA, Colburn NH . Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion. Oncogene 2010; 29: 3921–3932.
Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 2012; 40: W498–W504.
Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A . Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 2006; 174: 677–687.
Laird DW, Fistouris P, Batist G, Alpert L, Huynh HT, Carystinos GD et al. Deficiency of connexin43 gap junctions is an independent marker for breast tumors. Cancer Res 1999; 59: 4104–4110.
Qin H, Shao Q, Curtis H, Galipeau J, Belliveau DJ, Wang T et al. Retroviral delivery of connexin genes to human breast tumor cells inhibits in vivo tumor growth by a mechanism that is independent of significant gap junctional intercellular communication. J Biol Chem 2002; 277: 29132–29138.
Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005; 65: 5506–5511.
Shao Q, Wang H, McLachlan E, Veitch GI, Laird DW . Down-regulation of Cx43 by retroviral delivery of small interfering RNA promotes an aggressive breast cancer cell phenotype. Cancer Res 2005; 65: 2705–2711.
McLachlan E, Shao Q, Laird DW . Connexins and gap junctions in mammary gland development and breast cancer progression. J Membr Biol 2007; 218: 107–121.
Li Z, Zhou Z, Welch DR, Donahue HJ . Expressing connexin 43 in breast cancer cells reduces their metastasis to lungs. Clin Exp Metastasis 2008; 25: 893–901.
Plante I, Stewart MK, Barr K, Allan AL, Laird DW . Cx43 suppresses mammary tumor metastasis to the lung in a Cx43 mutant mouse model of human disease. Oncogene 2011; 30: 1681–1692.
Dick JE . Stem cell concepts renew cancer research. Blood 2008; 112: 4793–4807.
Sirnes S, Bruun J, Kolberg M, Kjenseth A, Lind GE, Svindland A et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int J Cancer 2012; 131: 570–581.
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131: 1109–1123.
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009; 138: 592–603.
Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011; 30: 2463–2474.
Deng L, Shang L, Bai S, Chen J, He X, Martin-Trevino R et al. MicroRNA100 inhibits self-renewal of breast cancer stem-like cells and breast tumor development. Cancer Res 2014; 74: 6648–6660.
McCarthy JJ . MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 2008; 1779: 682–691.
Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009; 326: 1549–1554.
Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab 2010; 12: 341–351.
Dey BK, Gagan J, Dutta A . miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 2011; 31: 203–214.
Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 2012; 122: 2054–2065.
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008; 451: 147–152.
Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D et al. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res 2011; 31: 3859–3863.
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y et al. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERalpha-positive endometrioid adenocarcinoma. Cancer Lett 2012; 314: 41–53.
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y et al. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun 2013; 433: 207–212.
Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010; 28: 341–347.
Kasinski AL, Slack FJ . Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011; 11: 849–864.
Kasinski AL, Slack FJ . Arresting the culprit: targeted antagomir delivery to sequester oncogenic miR-221 in HCC. Mol Ther Nucleic Acids 2012; 1: e12.
Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA 2012; 109: E1695–E1704.
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013; 368: 1685–1694.
Zuker M . Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31: 3406–3415.
Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D et al. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol 2011; 12: R77.
Lee CH, Kuo WH, Lin CC, Oyang YJ, Huang HC, Juan HF et al. MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. Int J Mol Sci 2013; 14: 11560–11606.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33: e179.
Acknowledgements
We thank Katri Selander, Gary Johnson and Jeffrey Green for providing cell lines. We thank Alana Welm for providing human mammary carcinoma xenografts. We thank Max Wicha for helpful discussions. This work was supported by NCI grant R01CA127405 (JMR), the Jo and Ben Statler Chair in Breast Cancer Research (JMR) and by grant R01CA148671 (ENP). Flow cytometry experiments were performed in the West Virginia University Flow Cytometry Core Facility, which is supported by NIH equipment grant RR020866/OD016165 and the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences under grants P30GM103488 (CoBRE) and P20GM103434 (INBRE). Orthotopic tumor cell injections were performed in the West Virginia University Animal Models and Imaging Facility, which has been supported by the Mary Babb Randolph Cancer Center and NIH grants P20RR016440, P30RR032138/GM103488 and S10RR026378.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Supplementary information
Rights and permissions
Oncogenesis is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lin, CC., Sharma, S., Farrugia, M. et al. Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis 4, e155 (2015). https://doi.org/10.1038/oncsis.2015.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2015.8
This article is cited by
-
circAKT3 positively regulates osteogenic differentiation of human dental pulp stromal cells via miR-206/CX43 axis
Stem Cell Research & Therapy (2020)
-
Connexin and pannexin channels in cancer
BMC Cell Biology (2016)
-
Regulation of the T-box transcription factor Tbx3 by the tumour suppressor microRNA-206 in breast cancer
British Journal of Cancer (2016)