Abstract
MicroRNAs (miRNAs) comprise a class of short, non-coding RNAs that regulate protein synthesis through posttranscriptional modifications. In this study, we found significant upregulation of miR-18a in prostate cancer specimens and prostate cancer cell lines compared with the normal controls. MiRNAs can be separated into two groups based on whether they regulate tumor suppressors or oncogenes. In our previous study, we found that miR-18a, which belongs to the miR17-92 cluster, is upregulated in prostate cancer; the objective of this study was to investigate the associated regulatory mechanisms. We found that miR-18a is upregulated in clinical tumor specimens and cancer cell lines. Our bioinformatics analysis showed that the serine/threonine-protein kinase 4 (STK4) 3′ untranslated region contains a highly conserved binding site for the miR-18a seed region. Luciferase reporter assays were performed to indicate that STK4 is a direct target of miR-18a. Interestingly, miR-18a knockdown decreased cell growth in prostate cancer cells and significantly decreased prostate tumor growth in in vivo nude mice experiments through STK4-mediated dephosphorylation of AKT and thereby inducing apoptosis. Our results suggest that miR-18a acts as an oncomiR targeting STK4 in prostate cancer, and inhibition of miR-18a expression may offer therapeutically beneficial option for prostate cancer treatment.
Similar content being viewed by others
Introduction
Prostate cancer is the fifth most commonly diagnosed cancer worldwide, and the incidence of prostate cancer has increased considerably over the past 25 years. In the United States, prostate cancer is the most commonly diagnosed solid cancer and the second leading cause of cancer deaths rate, following lung cancer.1 The incidence of prostate cancer increases with age, from 8.6% in men aged<45 years to 46% in menaged> 70 years.1 MicroRNAs (miRNAs) are short (approximately 22 nucleotides in length), single-stranded regulatory RNAs that interact with multiple mRNAs, resulting in their translational repression or degradation.2 Many miRNAs are differentially expressed in normal and tumor tissues and have biological functions in tumor progression. Certain miRNAs have been identified as biomarkers for cancers, such as ovarian, colon and pancreatic cancers. To date, a number of studies have identified approximately 100 miRNAs that are associated with the development of various cancers, including breast cancer, leukemia, lung cancer, colon cancer, brain cancer and liver cancer.3, 4
In recent years, miRNAs have been demonstrated to be involved in many diseases, including cancers. Therefore, studies of prostate cancer-specific miRNAs show potential for their utilization in the prevention, diagnosis and treatment of prostate cancer to more effectively target tumor growth and provide patients with better therapeutic options.5 Our previous miRNA microarray analysis revealed that three members of the miR-17-92 cluster, miR-17-3p, miR-17-5p and miR-18a, are upregulated in prostate cancer.6 A review of the existing literature shows that miR-18a has not been well studied in prostate cancer and that miRNAs belonging to the miR17-92 cluster are important oncomiRs in many tumors. The upregulation of the miR-17-92 cluster in various types of cancer implied an oncogenic role in the occurrence and progression of cancer.7, 8 However, the functional role and mechanism of action of miR-18a in prostate cancer have yet to be clarified. Recent studies also show an oncogenic role of miR-18a in breast cancer,9 nasopharyngeal cancer,10 pancreatic cancer11 and colorectal cancer,12 suggesting that miR-18a acts as an oncogenic miR in cancer progression. We hypothesize that miRNAs that are upregulated in cancers are likely to be oncomiRs that decrease the expression levels of tumor-suppressor proteins. In this study, we demonstrate that miR-18a exhibits an oncogenic role in prostate cancer by targeting serine/threonine-protein kinase 4 (STK4). STK4 has been identified as a tumor suppressor in colon cancer,13 soft tissue sarcoma14 and hepatic cell carcinoma.15, 16, 17 STK4, also known as STK4 (mammalian sterile 20-like kinase 1) and Krs-2,18, 19 is a 56–60-kDa protein that is highly conserved across different species. STK4 has a critical role in the apoptosis cascade and was found to have a crucial correlation with hepatocyte carcinoma development.15 Loss of STK4 protein expression has been observed in these different types of cancer. In addition, STK4 has been identified as a pro-apoptotic kinase and has acted as inhibitors of endogenous AKT.20 Thus, in this study, we aimed to investigate the role and function of miR-18a in regulation of STK4 in prostate cancer tumorigenesis.
Results
MiR-18a is highly expressed in prostate cancer tissues and prostate cancer cell lines
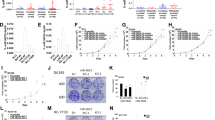
The miRNA microarray chip data indicate that miR-18a expression is higher in tumors than in normal tissues (Supplementary Figure S1). To confirm the miRNA microarray data, the expression of miR-18a was measured in 6 prostate cancer tissues and 10 benign prostatic hyperplasia tissues using quantitative reverse transcriptase-PCR (RT-PCR; Figure 1a). The expression level of miR-18a was significantly higher in prostate cancer tissue than in normal prostate tissue (P<0.01; Figure 1a) MiRNA expression was defined by an in situ hybridization (ISH) staining score. ISH of miR-18a was then performed to show that miR-18a is strongly expressed in most advanced prostate carcinomas compared with non-cancerous prostatic epithelium specimens (Figure 1b, left panel). Staining score was calculated as staining intensity multiplied by percentage of stained cells according to whole-cell ISH expression of miR-18a in non-tumor (NT) part as related to paired tumor tissues. Strikingly, we found that miR-18a levels were significantly upregulated in tumor part tissues compared with NT part tissues (P<0.005) (Figure 1b, bottom panel). To validate the expression level of miR-18a in prostate cancer tissues, we analyzed miR-18a expression levels from on-line publically available data set, deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE21032 and GSE21036.21 As shown in Figure 1c, miR-18a expression levels were significantly upregulated in prostate cancer tissues compared with the normal group (Figure 1c, right upper panel) (P<0.005). To further confirm the expression of miR-18a in NT paired tissues, five paired clinical samples were analyzed by miRNA-quantitative RT-PCR. As shown in Figure 1d, in five patients, miR-18a expression levels were higher in prostate cancer tissues compared with matched normal prostate tissues. The expression levels of miR-18a were also examined in prostate cell lines, showing that miR-18a were clearly upregulated in three human prostate cancer cell lines DU145, 22Rv-1 and LNCap compared with three immortalized prostate epithelium cell line PZH-HPV-7, PWR-1E and RWPE-1 (Figure 1e). Taken together, these results from clinical specimens and cell lines indicated that expression of miR-18a is significantly elevated in prostate cancer cells.
miR-18a is highly expressed in tumors. (a) The expression level of miR-18a in prostate cancer tissue. The differential expression of miR-18a in prostate cancer tissues was compared with that in normal tissues. The level of miR-18a was significantly higher in cancer tissues (P<0.01). (b) Left panel, ISH was used to detect the presence of miR-18a in non-tumor prostate and prostate cancer tissues. Representative ISH micrographs ( × 400) for miR-18a staining; right Panel, quantification according to ISH expression of miR-18a in prostate tumors as related to paired normal tissues. The scores are calculated as staining intensity × percentage of stained cells. (c) miR-18a relative expression level of non-tumor parts compared with tumor parts were analyzed using GEO data set. (d) The five NT paired tissues were used for examination of miR-18a expression. High expression of miR-18a tumor part specimens than that in non-tumor part tissues (e) The differential expression of miR-18a in prostate cancer cell lines was compared with that in prostate immortalized epithelium cell lines. The level of miR-18a was significantly higher in prostate cancer cell lines than in prostate immortalized epithelium cell lines.
STK4 is a candidate target gene of miR-18a
Computational analyses (TargetScan, PicTar, Mirnatargets and miRanda) identified 20 candidate proteins with 3′ untranslated regions (UTRs) containing potential miR-18a binding sites (Figure 2a). The top 20 candidate target proteins are listed in Supplementary Table S1. It is well known that decreased expression of tumor-suppressor miRs result in increased expression of target oncogenes, whereas increased expression of oncogenic miRs leads to loss or decreased expression of target tumor-suppressor genes. As shown in Figure 1, the expression level of miR-18a is upregulated in prostate cancer tissues; therefore, we hypothesized that tumor-suppressor genes, such as STK4, would be potential candidate target proteins. Other proteins identified as miR-18a targets in previous studies include SMAD2,22, 23 CTGF24 and CDC42. The miR-18a precursor was overexpressed in PC3 and DU45 prostate cancer cell lines to detect changes in the levels of candidate target proteins. Western blotting analysis was carried out at 72 h after miR-18a precursor transfection into PC-3 and DU145 cell lines, respectively. We found that STK4 protein expression decreased significantly in both PC-3 and DU145 cells overexpressing miR-18a, whereas the protein expression levels of SMAD2, CDC42 and CTGF in the miR-18a-overexpressing cells did not change significantly (Figure 2b). Next, we investigated whether there was an inverse correlation between miR-18a and STK4 protein in prostate cell lines and prostate clinical tissues. We examined the STK4 mRNA and protein expression levels in two immortalized prostate epithelium cell lines and four prostate cancer cell lines. The STK4 mRNA expression level was similar in all the six cell lines (Figure 2c, upper panel), and real-time PCR also demonstrated similar STK4 mRNA values in all the six cell lines (Supplementary Figure S2B). However, the STK4 protein level was higher in the immortalized cell lines, and its expression was inversely correlated with that of miR-18a (Figure 2c, bottom panel). These data showed that the STK4 protein level is regulated through posttranslational modification. In order to realize the STK4 protein expression of clinical tissues, we examined STK4 protein expression of clinical specimens by western blotting and immunohistochemistry (IHC) analysis. Western blotting indicated that the STK4 protein expression level was higher in the NT tissues than in the paired tumor tissues (Figure 2d upper panel). IHC analysis demonstrated strong cytosolic staining for STK4 in all NT prostate epithelium, whereas prostate adenocarcinomas showed no STK4 expression (Figure 2d bottom panel). The quantification of STK4 expression in paired prostate adenocarcinoma and corresponding normal tissue showed a significant downregulation in tumors compared with normal tissue (P<0.01; Figure 2d). We also used the GEO database to search the mRNA level of STK4 in NT paired prostate cancer. In the GEO data set (GDS2546/53858) indicated STK4 mRNA levels did not have the significant difference between NT parts paired with tumor tissues (P>0.05; Figure 2e). This data also confirm our experiment data, which indicates that STK4 mRNA levels are not different between normal and tumor tissues.
STK4 is inversely correlated with miR-18a. (a) Venn diagram displaying miRNAs computationally predicted to target STK4 by PicTar (red), TargetScan (green), miRanda (blue) and Mirnatargets (black). Examination of different databases to find candidate downstream target genes. (b) Protein expression of STK4, SMAD2, CDC42, CTGF and α-tubulin in control miR or miR-18a precursor transfected prostate cancer cells (PC-3, DU145). STK4 was suppressed by miR-18a transfection at the protein level in prostate cancer cell lines as shown by western blotting. (c) The expression of STK4 mRNA (upper panel) and protein (bottom panel) in six prostate cell lines was analyzed by RT-PCR and western blotting, respectively. The number below the GAPDH image was the density ratio of STK4/GAPDH (NIH-Image J) for mRNA (upper panel) and protein (bottom panel), respectively. The STK4 mRNA level was the same in 60 cell lines. The STK4 protein level is downregulated in four prostate cancer cell lines. (d) Western blotting (upper panel) and IHC (middle panel) analysis revealed STK4 protein level was downregulated in prostate tumor parts compared with adjacent non-tumor tissues. Quantification of STK4 expression in paired prostate adenocarcinoma and corresponding normal tissues (bottom panel) (n=10, P<0.001). Western blotting data showed that the STK4 protein is highly expressed in non-tumor compared with tumor tissues. IHC showed that STK4 staining is lost in the tumor tissues. (e) STK4 mRNA expression levels are not different between normal and tumor of prostate cancer in GEO data set validation. (f) ISH and IHC were used to detect the presence of miR-18a and STK4 in prostate cancer tissues. These sections of ISH and for IHC are consecutive sections. Quantitative IHC and ISH scores show that STK4 and miR-18a expression are inversely correlated. (g) miR-18a is highly expressed with poor tumor stage.
In prostate tumor tissues, prostate tumors with high endogenous miR-18a expression showed relatively low expression of STK4 protein (Figure 2f, upper panel, case 2), whereas the prostate tumors with low miR-18a expression showed relatively high levels of STK4 protein (Figure 2f, upper panel, case 1). The staining scores were obtained from immunoexpression of IHC and ISH and indicated inverse correlation between miR-18a and STK4 expression in prostate tumor tissues (Figure 2f, right panel, P<0.001). These results indicated that high miR-18a levels in prostate cancer cells played an oncogenic role by negatively regulating STK4 expression and that the downregulation of STK4 may be involved in prostate tumorigenesis. To address miR-18a expression level is correlated with which factor of clinical-pathology event. We used the prostate cancer tissue array obtained from BioMax Company (Rockville, MD, USA) to assay the correlation of miR-18a between TNM stage. We analyzed the clinical pathology of miR-18a expression level. The result showed that highly expressed miR-18a is correlated with poor stage of prostate cancer (Figure 2g, P<0.05).
MiR-18a can directly bind the STK4 3′UTR to suppress STK4 protein expression
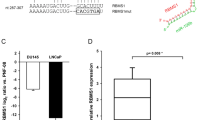
We next investigated whether the 3′UTR of STK4 was a functional target of miR-18a. We performed bioinformatics searches to find whether miR-18a may target nt 85–115 of 3′UTR of STK4. The schematic diagram of miR-18a binding sites in the 3′UTR of STK4 is shown in Figure 3a that was highly conserved across seven species. Because miR-18a has a predicted target site in the 3′UTR of STK4, we generated pMIR-reporter luciferase vectors containing the STK4 3′UTR sequence. In these vectors, the firefly luciferase coding sequence is followed by a 300 nucleotide PCR fragment encompassing the predicted miRNA binding site within the STK4 3′UTR, from nucleotides 17 to 317, as amplified from the wild type or seed region mutant constructs (Figure 3a, bottom panel, marked as Mut. 3′UTR). The recombinant STK4 3′UTR constructs were transfected into HEK-293T and DU-145 cells together with miR-18a or antagomiR-18a. The luciferase activity in HEK-293T and DU-145 cells transfected with STK4 wild type constructs plus miR-18a mimics was significantly lower than that in cells transfected with the control miR (Figure 3b, upper panel, lanes 2 and 3, P<0.05). The opposite result was observed in cells transfected with antagomiR-18a. In contrast, luciferase activities of mutant 3′UTR remained unchanged in miR-18a precursor overexpressing HEK293T cells (Figure 3c, lanes 4 and 5). The miR-18a expression level was examined by real-time PCR in all of the reporter assays (Supplementary Figures S3A and B). These results suggest that the STK4 mRNA is a direct target of miR-18a. To determine whether miR-18a inhibits endogenous STK4 expression, we used established human prostate cancer cell lines. As anticipated, the endogenous miR-18a levels were high in all the cells tested, making loss-of-function experiments challenging. The transfection efficiency of miR-18a precursor and anti-miR-18a in the cells has been examined by miRNA-quantitative RT-PCR (Supplementary Figures S3 A and B). We also examined the STK4 mRNA expression levels of reporter assay. In the real time PCR data showed STK4 mRNA levels did not change occur with miR-18a expression level (Supplementary Figures S3A and S3B). However, antagomiR-18a consistently and strongly increased STK4 protein and mRNA expression (Figure 3d and Supplementary Figure S3C). The selectivity was validated using a miRNA mimic approach. When the miR-18a mimic was transfected into RWPE-1 and PWR-1E cells, decreases in the STK4 protein and mRNA levels were observed (Figure 3d and Supplementary Figure S3C). The STK4 mRNA level alternation could be side effect. But the reporter assay showed that luciferase activities of mutant 3′UTR remained unchanged in miR-18a precursor overexpressing HEK293T cells (Figure 3c, lanes 4 and 5). These results indicated that miR-18a directly targets the STK4 3′UTR to suppress STK4 protein translation.
miR-18a directly binds the STK4 3′UTR to suppress the protein level. (a) Bioinformatics analysis demonstrates that the miR-18a seed region is highly conserved in different species. The computer-predicted binding energy is −21.1 Kcal/mol. (b) Luciferase reporter assay in HEK293T (upper panel) and DU145 (bottom panel) cells, with the cotransfection of WT-reporter and control-miR or pre-miR-18a as well as cotransfection of WT-reporter and control anti-miR or anti-miR-18a as indicated. Each bar represents the mean and s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001. (c) HEK-293T cells were transfected with the mutant STK4 3′UTR to examine the binding affinities of these sequences. The mutant UTRs of STK4 had no effect on reporter activity. (d) STK4 protein level after miR-18a knockdown in 22Rv1 and DU145 cell lines or overexpression of miR-18a in RWPE-1 and PWR-1E cell lines, respectively. At 48 h after transfection, total protein and RNA were extracted and used for western blotting (upper panel) and miRNA-quantitative RT-PCR (bottom panel) analysis.
MiR-18a enhances tumorigenicity by downregulating the STK4 protein in prostate cancer cell lines
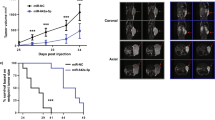
As overexpression of miR-18a implied an oncogenic role in prostate cancer, we decided to examine whether miR-18a has oncogenic functions in prostate cancer cells in vitro and in vivo. We analyzed clonogenic ability of prostate cancer cells by modulation of miR-18a or STK4 in both anchorage-dependent and anchorage-independent conditions. We found that the upregulation of miR-18a in PC3 increased colony formation by 50%, whereas the downregulation of miR-18a in 22Rv-1 and LNCap decreased colony formation by 30% (Figure 4a left panel, P<0.05, Figure 4b right panel, P<0.05 and Supplementary Figure S4A, P<0.001). The contrary effect was also observed in the green fluorescent protein (GFP)-STK4-overexpressing 22Rvl cells, which show that overexpression of STK4 significantly inhibited the proliferation rate of 22Rvl cells (Figure 4a, right panel, P<0.05). The same experiments were performed in another high endogenous miR-18a level prostate cancer cell line (Supplementary Figure S4A, P<0.001 in anti-miR-18a-transfected LNCap cells, P<0.05 in GFP-STK4-transfected LNCap cells). Interestingly, miR-18a suppression affected not only cell growth but also anchorage-independent growth. Transfection with an anti-miR-18a inhibitor and STK4 markedly decreased the plating efficiency of 22Rv-1 cells in soft agar (Figure 4c). Upregulation of miR-18a by the miR-18a precursor of PC-3 and DU145 cell lines and downregulation of miR-18a by the anti-miR-18a of 22Rv1 and LNCap cell lines were confirmed using miRNA-quantitative RT-PCR (Supplementary Figure S4B). Furthermore, we then investigated whether the oncogenic roles of miR-18a were dependent on STK4 expression. We transfected a STK4 construct into immortalized prostatic epithelial PWR-1E followed by transfection of the miR-18a precursor to perform colony-formation assay. In the colony-formation assay, PWR-1E cells transfected with a control miR and STK4 exhibited lower colony-formation ability than control miR and GFP-transfected cells. Conversely, cells transfected with pre-miR-18a and GFP exhibited an increased colony number compared with those transfected with pre-miR-18a and STK4. However, the effect of pre-miR-18a was not significantly different from that of STK4 (Figure 4d). These results supported the hypothesis that miR-18a executes an oncogenic effect on prostate cancer cells and the expression of miR-18a can inhibit prostate cancer cells’ growth in anchorage-dependent as well as -independent conditions. To determine whether miR-18a regulates tumor growth in vivo, we used tumor xenografts by inoculating 22Rv1 cells that had miR-18a suppressed in nude mice. Smaller subcutaneous tumors were observed in nude mice implanted with cells overexpressing antagomiR-18a and STK4 compared with those implanted with control cells (n=7–9/group; GFP compared with STK4 P<0.05, anti-NC compared with anti-miR-18a P<0.01, Figure 4e). At the end of the observation, the average volume (160±40 mm3) of tumors expressing anti-miR-18a was only about one-third of that (410±80 mm3) of the control group (Figure 4e, P<0.01). The similar result was also observed when 22Rv1 cells transfected with GFP-STK4 produced tumors much smaller than that of GFP control group (Figure 4e, right). At the end of the observation, the average tumor volume in the GFP-STK4 group was 110±20 mm3, whereas that in the GFP control group was 400±120 mm3, which represented a 3.0-fold decrease (P<0.05). These results indicate that miR-18a may act as an oncomiR in prostate cancer cells in vivo and in vitro.
miR-18a promotes the tumorigenicity of prostate cancer cells. (a) Decreased colony-formation ability is observed in 22Rv-1 cells overexpressing STK4 or antagomiR-18a. (b) PC3 cells expressing STK4 shRNA or pre-miR-18a exhibit increased colony-formation ability. (c) STK4 expression inhibits the oncogenic function of miR-18a. STK4 inhibits anchorage-independent growth of prostate cancer cells. 22Rv1 cells were transfected with STK4 or anti-miR-18a. Soft agar colony-formation assays were performed 24 h after transfection. (d) PWR-1E cells overexpressing pre-miR-18a or STK4 were subjected to a colony-formation assay to validate the effects of STK4 and miR-18a. (e) STK4 inhibits tumor growth of prostate cancer cells in nude mice. 22Rv1 cells were transfected with GFP-STK4, control GFP, anti-miR-18a or control anti-miR, respectively. Twenty-four hours after transfection, 1 × 106 cells were subcutaneously injected into of male nude mice. The dot plot show the tumor weight of each mice, and the inset photographs are representative xenografted tumors 6 weeks after inoculation. *P<0.05; **P<0.01. In vivo studies demonstrated that 22Rv-1 cells transfected with STK4 or antagomiR-18a are associated with decreased tumor volume and growth.
MiR-18a upregulation promotes tumor cell survival via suppression of the STK4-mediated AKT apoptosis cascade
Previous studies indicated that an increase in STK4 expression enhances apoptosis. The results described above indicated that inhibition of miR-18a expression or overexpressing of STK4 in the cells can inhibit the proliferation of 22Rvl cells (Figures 4a and b). Therefore, we examined the influence of miR-18a on cell proliferation using the MTT method to survey the number of live cells after the upregulation or downregulation of miR-18a expression. As expected, miR-18a downregulation suppressed the proliferation of prostate cancer cells. In contrast, STK4 upregulation significantly suppressed cell growth (Figure 5a). Previous studies indicated that STK4 is a pro-apoptotic kinase and is involved in the regulation of apoptosis.25, 26 We then performed cell cycle analysis by using flow cytometry to examine the role of STK4 on cell cycle distribution. We also examined the cell cycle distribution using flow cytometry. Flow cytometry data demonstrated that STK4 overexpression increased the subG1 fraction from 2% to 12% in a PC3 prostate cancer cell line (Supplementary Figure S5A). To further confirm cell death, we examined the expression of the apoptosis marker poly-(ADP-ribose) polymerase (PARP; which is hallmark of downstream apoptosis activation and cleaved during apoptosis) cleavage. Western blotting showed that prostate cancer cells transfected with antagomiR-18a exhibited increased levels of cleaved PARP (Figure 5b). Previous studies have reported that STK4 directly interacts with AKT to regulate AKT kinase activity. Other studies have also shown that AKT phosphorylation can prevent apoptosis via the phosphorylation of Caspase 9 and Bad. We then investigated whether STK4 induced cell apoptosis through the AKT-mediated pathway. As shown in Figure 5c, LNCap cells transfected with STK4 exhibited decreased AKT phosphorylation and increased PARP cleavage. This result strongly suggested that the dephosphorylation of AKT was involved in STK4-mediated apoptosis. We examined 22RV-1 cells overexpressing either STK4 to determine whether the observed anti-apoptotic effect was mediated by AKT regulation. The result of western blotting to detect phoshrylation of AKT s473 and PARP cleavage indicated the similar results with LNCap cell line. Furthermore, we used AKT inhibitor to examine whether AKT inhibitors synergize with STK4 in inducing apoptosis in 22Rv-1 cells. The results of the experiment are shown in Supplementary Figure S5C. After transfection of control vector and STK4 combined with myr-AKT for 24 h, the AKT inhibitor IV (1.25 μM, dissolved in dimethyl sulfoxide) was added into cells for 24 h. The synergistic effect of STK4 and AKT inhibitor IV-induced PARP cleavage were examined by western blotting analysis (Supplementary Figure S5C, compared lanes 3 and 4). These results indicate that STK4 induced prostate cancer cells apoptosis through STK4-mediated suppression of AKT phosphorylation, and reintroduction of STK4 protein may override the effects of miR-18a overexpression on the tumor growth of prostate cancer cells. Next, we performed IHC to examine protein expression level of STK4 and phosphorylation level of AKT in the subcutaneous tumor of the anti-miR-18a group and GFP-STK4 overexpression group of previous xenograft animal models. We also used IHC to examine STK4 expression and AKT phosphorylation in subcutaneous xenograft tissues. STK4 staining was observed in both the nucleus and cytosol in 22Rv-1 cells overexpressing STK4 and antago-miR-18a. Furthermore, 22Rv-1 cells overexpressing GFP and an antagomiR control showed elevated levels of phospho-AKT staining compared with cells overexpressing STK4 and antagomiR-18a. As shown, the expression levels of STK4 were inversely related to the phosphorylation levels of AKT in prostate cancer xenograft tumor tissues. We also used a human-specific antibody to confirm these expression patterns in xenograft tissue samples (Supplementary Figure S5B). The IHC results confirmed that STK4 expression is inversely correlated with AKT phosphorylation in subcutaneous xenograft tumors (Figure 5d). Taken together, these results indicated that miR-18a played an oncogenic role in the development of prostate cancer by downregulation of STK-mediated dephosphorylation of AKT signaling (Figure 5e).
miR-18a increased cell survival by suppressing STK4-mediated AKT kinase activity to inhibit apoptosis. (a) MTT assay indicating that the overexpression of STK4 and antagomiR-18a induces cell death in 22Rv-1 cells. (b) Western blotting of miRNA-transfected cells to detect the PARP cleavage level. (c) AKT phosphorylation in prostate cancer cells was suppressed by ectopic STK4 expression. Our observation in LNcap and 22Rv-1 prostate cancer cells transfected with STK4 suppressed AKT phosphorylation activity and increased PARP cleavage protein. (d) Representative images show that the expression levels of STK4 are inversely related to the phosphorylation levels of AKT in prostate cancer xenograft tumor tissues. (e) Diagram indicating how miR-18a regulates STK4 to act as an oncomiR in prostate cancer.
Discussion
We previously found that miR-18a acts as an oncomiR in prostate cancer. That study showed that miR-18a is highly expressed in prostate cancer clinical specimens using real-time PCR and ISH in prostate cancer cell lines. Using a database search, we determined that the STK4 3′UTR contains a candidate binding site for miR-18a. Western blotting and IHC demonstrated that STK4 and miR-18a protein expression are inversely correlated. In a reporter assay, we confirmed that the STK4 3′UTR is targeted by miR-18a. The overexpression of miR-18a in prostate cancer cells decreased the STK4 protein expression level and increased the colony-formation ability. Previous studies have indicated that STK4 can interact with AKT to regulate AKT kinase activity. Prostate cancer cells transfected with precursor miR-18a could decrease the STK4 protein level and enhance the AKT phosphorylation level. In vivo analysis also demonstrated that 22Rv-1 cells transfected with STK4 or antagomiR-18a suppressed tumor growth (decreased tumor volume). The STK4 expression level in clinical prostate cancer specimens, as measured by IHC, was higher in NT than tumor tissues. To the best of our knowledge, this study is the first to report that inhibition of miR-18a offer a therapeutically beneficial option for prostate cancer treatment, as evidenced by studies using clinical sample, cell lines and animal models and analyses.
MiR-18a belongs to the miR-17-92 cluster. Previous studies have confirmed that the 17–92 cluster is highly expressed in several types of cancers.11, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36 MiR-18a was expressed as part of the miR-17-92 cluster, which consists of seven miRNAs: miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a. This cluster has been reported to have a potential oncogenic function in various tumors.11, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36 In our microarray, we found that, in addition to miR-18a, miR-17-5p and miR-17-3p are also highly expressed in tumors compared with benign prostatic hyperplasia specimens. One report has also shown that miR-17-5p is upregulated in prostate cancer tissues.37 But, in our miRNA-quantitative RT-PCR, result has shown that the expression pattern of miR-17-5p was not similar with miR-18a in prostate cancer cell lines (Supplementary Figure S2A). Therefore, we selected miR-18a as a new candidate miRNA of miR-17-92 cluster to have an oncogenic role in prostate cancer tumorigenesis. Thus, no 17-92 cluster miRNAs other than miR18a is likely to be involved in prostate cancer progression.38, 39, 40
We next used several algorithms to identify potential target tumor-suppressor genes of miR-18a. The prediction results of integrated four software programs identified that the 3′-UTR of STK4 mRNA has putative miR-18a binding site. To understand the correlation between miR-18a and STK4, we performed IHC and ISH analysis and observed an inverse correlation of miR-18a and STK4 protein expression. We also successfully verified that STK4 is negatively regulated by miR-18a at the posttranscriptional level via a specific target site (nt 85–115) within the 3′UTR. As shown with 3′UTR luciferase assay and western blotting analyses, the STK4 gene expression was regulated by miR-18a. Collectively, these results showed that the expression of STK4 was regulated by miR-18a and that the level of STK4 protein expression was inversely correlated with miR-18a expression in prostate cancer cells.
The polycistronic miRNA cluster termed miR-17-92, which is located on chromosome 13 open reading frame 25 in the human genome, encodes seven miRNAs: miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92-1. The amplification of 13q31-q32, which is the locus containing the miR-17-92 cluster, has been reported in other solid tumors. A previous study had indicated that E2F and miR-20 participate in an autoregulatory feedback loop that regulates miR-17 and miR-20 expression.41 However, the present study did not reveal the mechanism that regulates miR-18a expression.
MiR-17-3p has been demonstrated to enhance the tumorigenicity of prostate cancer.37 In addition, it was shown that the increase in the miR-17-92 cluster expression level is due to a DNA copy number gain in the miR-17-92 cluster locus at 13q31. Other studies have indicated that the miR-17-92 cluster promotes oncogenesis in other solid tumors, but we found that miR-18a only serves as an oncomiR in prostate cancer progression. In the clinical pathology analysis, result demonstrated that highly miR-18a expression are correlated with poor tumor stage. Kaplan-Merrier curve indicated that upregulated miR-18a is correlated with poor survival, but the case numbers are not enough for power (data not shown).
STK4, a homolog of Hippo (Hpo/hpo) in Drosophila, was originally identified as a proapoptotic protein.42, 43 In addition to their proapoptotic function, STK4 has been shown to have an important role in cell-cycle progression and tumorigenesis. We also found that STK4 interacts with AKT to regulate its kinase activity. AKT is a known apoptosis inhibitor that can phosphorylate Caspase 9 and Bad to suppress the apoptosis cascade. Another study showed that STK4 can induce and enhance apoptosis. Our study indicated that STK4 could regulate apoptosis in prostate cancer cell lines by regulating AKT kinase activity. One study also indicated that miR-18a could act as an oncomiR to promote tumor cell proliferation and a malignant phenotype in HCC.44 The miR-17-92 cluster is highly expressed in gastric tumors. Our data show that the upregulation of miR-18a binds directly to the 3′UTR of the STK4 mRNA, downregulating STK4 at the protein level and thus suppressing apoptosis and promoting tumor survival. A previous study demonstrated that the miR-17-92 cluster is associated with 13q gain during the transformation of adenoma into adenocarcinoma during colon cancer progression.31 However, our data do not address whether miR-18a is involved in prostate cancer progression.
Prostate cancer is the second most common cause of death in Western countries. Evidence has shown that STK4 acts as a tumor suppressor in prostate cancer. Based on our present findings, we propose that miR-18a directly binds the STK4 3′UTR to suppress STK4 protein translation, leading to decreased interaction with AKT and ultimately promoting tumor cell survival. The implication from our data is that the upregulation of miR-18a induces a decrease in the STK4 protein level and an increase in AKT phosphorylation to enhance tumor cell survival. These data, together with our data from resected patient prostate tumors, suggests that targeting the miR-18a/STK4 interaction or rescuing STK4 expression may prove to be a new therapeutic strategy in the treatment of prostate cancer patients. Therefore, the inhibition of miR-18a may also be a therapy target in the future.
Materials and methods
IHC
Tissue section microarray blocks were transferred to coated slides. Ten prostate cancer samples were immunostained with an antibody to the N-terminal region of STK4/STK4 (Abcam, Cambridge, MA, USA). The slides were dewaxed and rehydrated in dH2O, and sections were immersed in citrate buffer (pH 6.0) and then microwaved for 20 min for antigen retrieval. Endogenous peroxidase activity was blocked using 0.5% H2O2. The slides were then transferred into a humidified chamber, incubated with 5% horse serum for 30 min and then incubated with STK4/STK4 primary antibody overnight at 4 °C. After primary antibody incubation, the slides were immersed in peroxidase-labeled secondary antibody for 30 min at room temperature. To detect the antibody-conjugated antigen reaction, the sections were incubated in 3-amino-9-ethylcarbazole substrate-chromogen for 30 min and counterstained with hematoxylin (K4005, EnVision System-HRP (AEC); DakoCytomation, Carpinteria, CA, USA).
Statistical analysis
All tests were two-sided, and P<0.05 was accepted statistically significant. SPSS17 software (New York, NY, USA) was used to analyze all clinical parameters.
MiRNA detection
To determine whether miRNA candidates were involved in STK4 regulation and served as putative tumor suppressors in prostate cancer, we first examined the expression levels of candidate miRNAs in prostate cancer cell lines and 10 tumor specimens by miR-quantitative RT–PCR analysis. Standard protocols were followed for total RNA extraction and reverse transcription. RNA and cell lysate samples were prepared from human prostate cancer cell lines and human immortalized normal prostate epithelium cell lines.
Reverse transcription reactions for the cell lines and human prostate cancer specimens were performed in reactions containing 10 ng total RNA. The amplification and detection of specific products was performed with a Roche LightCycler detection system with the cycling profile specified in the TaqMan qRT–PCR miRNA Detection Kit (Ambion, New York, NY, USA). Relative gene expression was calculated by comparing the cycle times for each target PCR. The target PCR Ct values were normalized by subtracting the Ct value of the internal control snoRNA RNU6B.
Reporter assay
The STK4 3′UTR target site was amplified by PCR and cloned downstream of the luciferase gene in the pMIR-REPORT luciferase vector (Ambion). This vector was sequenced and named STK4-3′UTR. For reporter assays, the cells were transiently transfected with the ST4-3′UTR plasmid, an artificial miRNA precursor and the Renilla luciferase plasmid as a normalization control using Lipofectamine 2000 (Invitrogen, New York, NY, USA). Reporter assays were performed 48 h posttransfection using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA).
Colony-formation assay
For the colony-formation assay, cells seeded in 60-mm dishes at 60–80% confluence were transfected with Lipofectamine 2000 and 30 nM of miR-NC or miRNA-18a oligonucleotide. After 24 h, the cells were trypsinized, split and plated at 500 cells/well in a six-well plate containing RPMI/10% fetal bovine serum (FBS) medium. After 7–14 days, the cells were fixed with formaldehyde and stained with crystal violet. A scanner was used to import data from each plate into a computer, and the results were analyzed with ImageJ (Maryland, MD, USA).
Soft agar assay
The soft agar colony-formation assay was performed to further assay the ability of cells to grow in suspension in agar (that is, without attachment to a surface). For the soft agar assay, cells were seeded in 35-mm dishes at 60–80% confluence and transfected with 30 nM of antagomiR-NC, antago-miRNA oligonucleotide or GFP and GFP-STK4 using Lipofectamine 2000. Agarose (0.5%) containing RPMI/10% FBS medium was added to a six-well plate as a lower layer. Twenty-four hours later, the cells were trypsinized and split in 0.35% agarose containing RPMI/10% FBS medium; 5000 cells/well were plated in six-well dishes.
Cell culture
The 22Rv-1, PC3 and LNCap cell lines (ATCC, Rockville, MD, USA) were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin and streptomycin. DU-145 cells were cultured in Minimum Essential Medium containing 10% FBS and antibiotics. Cultures were maintained in a 5% CO2 atmosphere at 1 atmosphere in a humidified incubator at 37 °C.
Western blotting
Lysis buffer was added to the cell lines for 30 min, and lysates were collected with cell scrapers at 4 °C. Sample buffer was added to the lysates, which were heated to 99 °C for 10 min. The protein samples were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the proteins were transferred to a polyvinylidene difluoride membrane using an immersion transfer device. After blocking in 5% milk in TBST (Tris-buffered saline with Tween 20), the membrane was incubated with primary antibody overnight at 4 °C. Membranes were washed three times with TBST for 10 min, and the membranes were incubated with appropriate secondary antibodies. X-ray films were used to detect the horseradish peroxidase signal.
ISH
Clinical specimens were infiltrated with 10% formalin solution and embedded in paraffin. The sections were deparaffinized, rehydrated through immersion in xylene and ethanol and then fixed with 4% paraformaldehyde (Sigma-Aldrich, St Louis, MO, USA) in diethylpyrocarbonate-phosphate-buffered saline at room temperature for 20 min. The sections were treated with acetylation solution for 10 min followed by phosphate-buffered saline containing 10 μg/ml proteinase K (Biochain, Newark, CA, USA) for 10 min at 37 °C. After this step, the slides were washed once and again incubated in 4% paraformaldehyde (Sigma-Aldrich) in diethylpyrocarbonate -phosphate-buffered saline for fixation. The sections were then blocked with hybridization solution for 4 h at room temperature and incubated with digoxigenin (Dig)-conjugated hsa-miR-18a probes (Exiqon, Vedbaek, Denmark) or Dig-conjugated control probes with a scrambled sequence (Exiqon) overnight at 45 °C. The slides were washed with saline-sodium citrate buffer and then incubated with blocking solution for 1 h at room temperature. The sections were incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody (Biochain) overnight at 4 °C. After two washes with alkaline phosphatase buffer, the sections were developed with NBT/BCIP (Biochain) for 24 h, with light blue cytoplasmic staining considered positive. Finally, the sections were counterstained with Nuclear Fast Red.
Clinical specimens
Human prostate clinical specimens were obtained from Kaohsiung Veterans General Hospital (Kaohsiung, Taiwan) and National Cheng Kung University Hospital (Tainan, Taiwan). All patients were informed of their rights and consented to the study. The specimen and patient data collection was performed following the protocol approved by the IRB Committee before tissue sample collection during the planned surgery. Carcinoma samples were obtained from palliative transurethral excisions of the prostate, and hematoxylin and eosin-stained tumor tissue sections were examined by a pathologist. Benign samples were obtained from transurethral resections of the prostate from patients treated for BPH. The specimens were histologically verified not to contain any prostate cancer cells. These samples were snap frozen and stored in liquid nitrogen until use. The prostate cancer tissue were obtained from BioMax company.
References
Jemal A, Siegel R, Xu J, Ward E . Cancer statistics, 2010. CA Cancer J Clin 2010; 60: 277–300.
Le Quesne JP, Spriggs KA, Bushell M, Willis AE . Dysregulation of protein synthesis and disease. J Pathol 2010; 220: 140–151.
Visone R, Petrocca F, Croce CM . Micro-RNAs in gastrointestinal and liver disease. Gastroenterology 2008; 135: 1866–1869.
Murray MY, Rushworth SA, MacEwan DJ . Micro RNAs as a new therapeutic target towards leukaemia signalling. Cell Signal 2012; 24: 363–368.
Deng JH, Deng Q, Kuo CH, Delaney SW, Ying SY . MiRNA targets of prostate cancer. Methods Mol Biol 2013; 936: 357–369.
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG et al. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009; 28: 3360–3370.
Yu G, Tang JQ, Tian ML, Li H, Wang X, Wu T et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol 2012; 106: 232–237.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005; 65: 9628–9632.
Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA 2009; 106: 15732–15737.
Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang J et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2013; 34: 415–425.
Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer 2011; 105: 1733–1740.
Vega AB, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep 2013; 30: 320–326.
Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR et al. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod Pathol 2007; 20: 331–338.
Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog 2007; 46: 865–871.
Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009; 16: 425–438.
Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA 2010; 107: 1437–1442.
Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA 2010; 107: 1431–1436.
Creasy CL, Ambrose DM, Chernoff J . The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J Biol Chem 1996; 271: 21049–21053.
Taylor LK, Wang HC, Erikson RL . Newly identified stress-responsive protein kinases, Krs-1 and Krs-2. Proc Natl Acad Sci USA 1996; 93: 10099–10104.
Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N et al. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J 2007; 26: 4523–4534.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Dogar AM, Towbin H, Hall J . Suppression of latent transforming growth factor (TGF)-beta1 restores growth inhibitory TGF-beta signaling through microRNAs. J Biol Chem 2011; 286: 16447–16458.
Li L, Shi JY, Zhu GQ, Shi B . MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem 2012; 113: 1235–1244.
Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A . Targeting of TGFbeta signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA 2013; 19: 177–190.
Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 2003; 113: 507–517.
Ura S, Masuyama N, Graves JD, Gotoh Y . Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proc Natl Acad Sci USA 2001; 98: 10148–10153.
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009; 24: 652–657.
Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 2009; 34: 1069–1075.
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L et al. MicroRNA profiling of human gastric cancer. Mol Med Rep 2009; 2: 963–970.
Zhang ZW, An Y, Teng CB . The roles of miR-17-92 cluster in mammal development and tumorigenesis. Yi Chuan 2009; 31: 1094–1100.
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ . Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis 2010; 11: 50–54.
Yeh SH, Chen PJ . Gender disparity of hepatocellular carcinoma: the roles of sex hormones. Oncology 2010; 78 (Suppl 1): 172–179.
Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res 2011; 17: 4125–4135.
Brock M, Trenkmann M, Gay RE, Gay S, Speich R, Huber LC . MicroRNA-18a enhances the interleukin-6-mediated production of the acute-phase proteins fibrinogen and haptoglobin in human hepatocytes. J Biol Chem 2011; 286: 40142–40150.
Yang Y, Ding L, An Y, Zhang ZW, Lang Y, Tai S et al. MiR-18a regulates expression of the pancreatic transcription factor Ptf1a in pancreatic progenitor and acinar cells. FEBS Lett 2012; 586: 422–427.
Wu W, Takanashi M, Borjigin N, Ohno SI, Fujita K, Hoshino S et al. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br J Cancer 2013; 108: 653–661.
Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res 2013; 41: 9688–9704.
Sikand K, Slane SD, Shukla GC . Intrinsic expression of host genes and intronic miRNAs in prostate carcinoma cells. Cancer Cell Int 2009; 9: 21.
Xu Y, Fang F, Zhang J, Josson S St, Clair WH St, Clair DK . miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One 2010; 5: e14356.
Kim K, Chadalapaka G, Pathi SS, Jin UH, Lee JS, Park YY et al. Induction of the transcriptional repressor ZBTB4 in prostate cancer cells by drug-induced targeting of microRNA-17-92/106b-25 clusters. Mol Cancer Ther 2012; 11: 1852–1862.
Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 2007; 282: 2135–2143.
Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S . MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J Biol Chem 2001; 276: 19276–19285.
Reszka AA, Halasy-Nagy JM, Masarachia PJ, Rodan GA . Bisphosphonates act directly on the osteoclast to induce caspase cleavage of mst1 kinase during apoptosis. A link between inhibition of the mevalonate pathway and regulation of an apoptosis-promoting kinase. J Biol Chem 1999; 274: 34967–34973.
Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 2009; 136: 683–693.
Acknowledgements
We thank the National Science Council (NSC 102-2325-B-006-011 and DOH102-TD-PB-111-TM027 to P-JL) for the grant support. We thank the proteomics core facility of the Clinical Medicine Research Center in National Cheng Kung University Hospital for assistance with protein experiment processing.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Hsu, TI., Hsu, CH., Lee, KH. et al. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 3, e99 (2014). https://doi.org/10.1038/oncsis.2014.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2014.12
Keywords
This article is cited by
-
Inhibiting anti-angiogenic VEGF165b activates a miR-17-20a-Calcipressin-3 pathway that revascularizes ischemic muscle in peripheral artery disease
Communications Medicine (2024)
-
MiR-211 determines brain metastasis specificity through SOX11/NGN2 axis in triple-negative breast cancer
Oncogene (2021)
-
MicroRNA mediated therapeutic effects of natural agents in prostate cancer
Molecular Biology Reports (2021)
-
The dual functional role of MicroRNA‐18a (miR‐18a) in cancer development
Clinical and Translational Medicine (2019)
-
SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2
Cell Death & Differentiation (2019)