Abstract
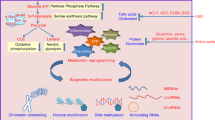
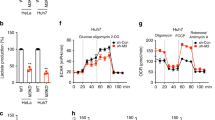
The anti-diabetic biguanide metformin may exert health-promoting effects via metabolic regulation of the epigenome. Here we show that metformin promotes global DNA methylation in non-cancerous, cancer-prone and metastatic cancer cells by decreasing S-adenosylhomocysteine (SAH), a strong feedback inhibitor of S-adenosylmethionine (SAM)-dependent DNA methyltransferases, while promoting the accumulation of SAM, the universal methyl donor for cellular methylation. Using metformin and a mitochondria/complex I (mCI)-targeted analog of metformin (norMitoMet) in experimental pairs of wild-type and AMP-activated protein kinase (AMPK)-, serine hydroxymethyltransferase 2 (SHMT2)- and mCI-null cells, we provide evidence that metformin increases the SAM:SAH ratio-related methylation capacity by targeting the coupling between serine mitochondrial one-carbon flux and CI activity. By increasing the contribution of one-carbon units to the SAM from folate stores while decreasing SAH in response to AMPK-sensed energetic crisis, metformin can operate as a metabolo-epigenetic regulator capable of reprogramming one of the key conduits linking cellular metabolism to the DNA methylation machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA . Metformin as a Tool to target aging. Cell Metab 2016; 23: 1060–1065.
López-Otín C, Galluzzi L, Freije JM, Madeo F, Kroemer G . Metabolic control of longevity. Cell 2016; 166: 802–821.
Kinnaird A, Zhao S, Wellen KE, Michelakis ED . Metabolic control of epigenetics in cancer. Nat Rev Cancer 2016; 16: 694–707.
Sharma U, Rando OJ . Metabolic inputs into the epigenome. Cell Metab 2017; 25: 544–558.
Zhong T, Men Y, Lu L, Geng T, Zhou J, Mitsuhashi A et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2017; 36: 2345–2354.
Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun 2015; 6: 10221.
Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr 2001; 131: 2811–2818.
Owen MR, Doran E, Halestrap AP . Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 2000; 348: 607–614.
Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife 2014; 3: e02242.
Konishi H, Mohseni M, Tamaki A, Garay JP, Croessmann S, Karnan S et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc Natl Acad Sci USA 2011; 108: 17773–17778.
Menendez JA, Folguera-Blasco N, Cuyàs E, Fernández-Arroyo S, Joven J, Alarcón T . Accelerated geroncogenesis in hereditary breast-ovarian cancer syndrome. Oncotarget 2016; 7: 11959–11971.
Cuyàs E, Fernández-Arroyo S, Alarcón T, Lupu R, Joven J, Menendez JA . Germline BRCA1 mutation reprograms breast epithelial cell metabolism towards mitochondrial-dependent biosynthesis: evidence for metformin-based ‘starvation’ strategies in BRCA1 carriers. Oncotarget 2016; 7: 52974–52992.
Sedic M, Skibinski A, Brown N, Gallardo M, Mulligan P, Martinez P et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat Commun 2015; 6: 7505.
Fernández-Arroyo S, Cuyàs E, Bosch-Barrera J, Alarcón T, Joven J, Menendez JA . Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience 2016; 2: 958–967.
De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013; 12: 247–256.
Miousse IR, Koturbash I . The Fine LINE: methylation drawing the cancer landscape. Biomed Res Int 2015; 2015: 131547.
Kitkumthorn N, Mutirangura A . Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet 2011; 2: 315–330.
Pal S, Tyler JK . Epigenetics and aging. Sci Adv 2016; 2: e1600584.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009; 138: 645–659.
Vazquez-Martin A, Oliveras-Ferraros C, Cufí S, Del Barco S, Martin-Castillo B, Menendez JA . Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell Cycle 2010; 9: 3807–3814.
Cuyàs E, Corominas-Faja B, Menendez JA . The nutritional phenome of EMT-induced cancer stem-like cells. Oncotarget 2014; 5: 3970–3982.
Banerjee P, Surendran H, Chowdhury DR, Prabhakar K, Pal R . Metformin mediated reversal of epithelial to mesenchymal transition is triggered by epigenetic changes in E-cadherin promoter. J Mol Med (Berl) 2016; 94: 1397–1409.
Yang M, Soga T, Pollard PJ . Oncometabolites: linking altered metabolism with cancer. J Clin Invest 2013; 123: 3652–3658.
Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, Powell DR et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res 2012; 22: 2339–2355.
Menendez JA, Corominas-Faja B, Cuyàs E, García MG, Fernández-Arroyo S, Fernández AF et al. Oncometabolic nuclear reprogramming of cancer stemness. Stem Cell Rep 2016; 6: 273–283.
Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F . Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012; 122: 253–270.
Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B . Metformin: from mechanisms of action to therapies. Cell Metab 2014; 20: 953–966.
Pollak M . Potential applications for biguanides in oncology. J Clin Invest 2013; 123: 3693–3700.
Boukalova S, Stursa J, Werner L, Ezrova Z, Cerny J, Bezawork-Geleta A et al. Mitochondrial targeting of metformin enhances its activity against pancreatic cancer. Mol Cancer Ther 2016; 15: 2875–2886.
Bridges HR, Sirviö VA, Agip AN, Hirst J . Molecular features of biguanides required for targeting of mitochondrial respiratory complex I and activation of AMP-kinase. BMC Biol 2016; 14: 65.
Mattaini KR, Sullivan MR, Vander Heiden MG . The importance of serine metabolism in cancer. J Cell Biol 2016; 214: 249–257.
Meiser J, Vazquez A . Give it or take it: the flux of one-carbon in cancer cells. FEBS J 2016; 283: 3695–3704.
Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N et al. Serine one-carbon catabolism with formate overflow. Sci Adv 2016; 2: e1601273.
Locasale JW . Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 2013; 13: 572–583.
Kotecki M, Reddy PS, Cochran BH . Isolation and characterization of a near-haploid human cell line. Exp Cell Res 1999; 252: 273–280.
Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res 2014; 24: 2059–2065.
Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 2015; 21: 81–94.
Cheng G, Zielonka J, Ouari O, Lopez M, McAllister D, Boyle K et al. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res 2016; 76: 3904–1395.
Ulanovskaya OA, Zuhl AM, Cravatt BF . NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol 2013; 9: 300–306.
Yang Q, Liang X, Sun X, Zhang L, Fu X, Rogers CJ et al. AMPK/α-ketoglutarate axis dynamically mediates DNA demethylation in the Prdm16 promoter and brown adipogenesis. Cell Metab 2016; 24: 542–554.
Kodiha M, Ho-Wo-Cheong D, Stochaj U . Pharmacological AMP-kinase activators have compartment-specific effects on cell physiology. Am J Physiol Cell Physiol 2011; 301: C1307–C1315.
Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ . Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 2013; 494: 256–260.
Talarico G, Orecchioni S, Dallaglio K, Reggiani F, Mancuso P, Calleri A et al. Aspirin and atenolol enhance metformin activity against breast cancer by targeting both neoplastic and microenvironment cells. Sci Rep 2016; 6: 18673.
Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL et al. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J 2008; 412: 211–221.
Houde VP, Ritorto MS, Gourlay R, Varghese J, Davies P, Shpiro N et al. Investigation of LKB1 Ser431 phosphorylation and Cys433 farnesylation using mouse knockin analysis reveals an unexpected role of prenylation in regulating AMPK activity. Biochem J 2014; 458: 41–56.
Ross FA, Jensen TE, Hardie DG . Differential regulation by AMP and ADP of AMPK complexes containing different γ subunitisoforms. Biochem J 2016; 473: 189–199.
Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M et al. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res 2014; 74: 7521–7533.
Liu X, Romero IL, Litchfield LM, Lengyel E, Locasale JW . Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab 2016; 24: 728–739.
Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 2017; 544: 372–376.
Corominas-Faja B, Quirantes-Piné R, Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Martin-Castillo B et al. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany NY) 2012; 4: 480–498.
Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cochemé HM, Noori T et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 2013; 153: 228–239.
Newman AC, Maddocks ODK . One-carbon metabolism in cancer. Br J Cancer 2017; 116: 1499–1504.
Newman AC, Maddocks ODK . Serine and functional metabolites in cancer. Trends Cell Biol 2017; 27: 645–657.
Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J et al. 5'-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol 2006; 26: 5336–5347.
Iglesias T, Espina M, Montes-Bayón M, Sierra LM, Blanco-González E . Anion exchange chromatography for the determination of 5-methyl-2'-deoxycytidine: application to cisplatin-sensitive and cisplatin-resistant ovarian cancer cell lines. Anal Bioanal Chem 2015; 407: 2423–2431.
Acknowledgements
This work was supported in part by grants from the Ministerio de Ciencia e Innovación (Grant SAF2016-80639-P), Plan Nacional de I+D+I, Spain and the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) (Grant 2014 SGR229), Department d’Economia I Coneixement, Catalonia, Spain, to Javier A Menendez and grants from the Czech Science Foundation (16-12816S) to Jan Stursa and Lukas Werner and the Czech Health Research Council (16-31704A) to Jiri Neuzil. Elisabet Cuyàs is supported by a Sara Borrell post-doctoral contract CD15/00033 from the Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria (FIS), Spain, The Metabolism and Cancer laboratory is supported by an unrestricted grant from the Armangué family (Girona, Catalonia). This work is in memory of Joan Armangué who passed away after his brave fight against cancer in November 2016.
Author contributions
JAM conceived the idea, directed the project and wrote the manuscript. EC, SF-A, SV, RA-FG, MM-B and EB-G conducted metabolomic, ELISA-/HPLC-based DNA methylation and western blot experiments, and analyzed the data. JS and LW provided essential reagents. JN and BV provided essential reagents, intellectual insights and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Cuyàs, E., Fernández-Arroyo, S., Verdura, S. et al. Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene 37, 963–970 (2018). https://doi.org/10.1038/onc.2017.367
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.367
This article is cited by
-
Metformin and cancer hallmarks: shedding new lights on therapeutic repurposing
Journal of Translational Medicine (2023)
-
Underexplored reciprocity between genome-wide methylation status and long non-coding RNA expression reflected in breast cancer research: potential impacts for the disease management in the framework of 3P medicine
EPMA Journal (2023)
-
Elevated RIF1 participates in the epigenetic abnormalities of zygotes by regulating histone modifications on MuERV-L in obese mice
Molecular Medicine (2022)
-
Repurposing dextromethorphan and metformin for treating nicotine-induced cancer by directly targeting CHRNA7 to inhibit JAK2/STAT3/SOX2 signaling
Oncogene (2021)
-
Serine hydroxymethyltransferase 2: a novel target for human cancer therapy
Investigational New Drugs (2021)