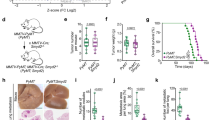
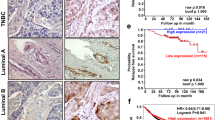
Abstract
Recently, we reported that the histone methyltransferase, EZH2, controls leukocyte migration through interaction with the cytoskeleton remodeling effector, VAV, and direct methylation of the cytoskeletal regulatory protein, Talin. However, it is unclear whether this extranuclear, epigenetic-independent function of EZH2 has a profound impact on the initiation of cellular transformation and metastasis. Here, we show that EZH2 increases Talin1 methylation and cleavage, thereby enhancing adhesion turnover and promoting accelerated tumorigenesis. This transforming capacity is abolished by targeted disruption of EZH2 interaction with VAV. Furthermore, our studies demonstrate that EZH2 in the cytoplasm is closely associated with cancer stem cell properties, and that overexpression of EZH2, a mutant EZH2 lacking its nuclear localization signal (EZH2ΔNLS), or a methyl-mimicking Talin1 mutant substantially promotes JAK2-dependent STAT3 activation and cellular transformation. Taken together, our results suggest a critical role for the VAV interaction-dependent, extranuclear action of EZH2 in neoplastic transformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611.
Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010; 42: 181–185.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008; 27: 7274–7284.
Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell 2011; 19: 86–100.
Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res 2012; 72: 3091–3104.
Yoo KH, Hennighausen L . EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci 2012; 8: 59–65.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–874.
Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43: 798–810.
Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA 2014; 111: 3098–3103.
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338: 1465–1469.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013; 23: 839–852.
Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol 2016; 23: 631–639.
He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 2012; 26: 37–42.
Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell 2012; 48: 572–586.
Hock H . A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev 2012; 26: 751–755.
Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 2012; 18: 298–301.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Chou RH, Yu YL, Hung MC . The roles of EZH2 in cell lineage commitment. Am J Transl Res 2011; 3: 243–250.
Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009; 136: 1122–1135.
Hobert O, Jallal B, Ullrich A . Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol 1996; 16: 3066–3073.
Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 2005; 121: 425–436.
Su IH, Tarakhovsky A . Lysine methylation and 'signaling memory'. Curr Opin Immunol 2006; 18: 152–157.
Boddicker RL, Razidlo GL, Dasari S, Zeng Y, Hu G, Knudson RA et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood 2016; 128: 1234–1245.
Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48: 607–616.
Chang KH, Sanchez-Aguilera A, Shen S, Sengupta A, Madhu MN, Ficker AM et al. Vav3 collaborates with p190-BCR-ABL in lymphoid progenitor leukemogenesis, proliferation, and survival. Blood 2012; 120: 800–811.
Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015; 47: 1304–1315.
Menacho-Marquez M, Garcia-Escudero R, Ojeda V, Abad A, Delgado P, Costa C et al. The Rho exchange factors Vav2 and Vav3 favor skin tumor initiation and promotion by engaging extracellular signaling loops. PLoS Biol 2013; 11: e1001615.
Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F et al. The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal 2012; 5: ra71.
Thalappilly S, Soubeyran P, Iovanna JL, Dusetti NJ . VAV2 regulates epidermal growth factor receptor endocytosis and degradation. Oncogene 2010; 29: 2528–2539.
Aghazadeh B, Lowry WE, Huang XY, Rosen MK . Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 2000; 102: 625–633.
Katzav S, Martin-Zanca D, Barbacid M . vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J 1989; 8: 2283–2290.
Groysman M, Nagano M, Shaanan B, Katzav S . Mutagenic analysis of Vav reveals that an intact SH3 domain is required for transformation. Oncogene 1998; 17: 1597–1606.
Hobert O, Sures I, Ciossek T, Fuchs M, Ullrich A . Isolation and developmental expression analysis of Enx-1, a novel mouse Polycomb group gene. Mech Dev 1996; 55: 171–184.
Rudolph MG, Stanfield RL, Wilson IA . How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol 2006; 24: 419–466.
Jiao L, Liu X . Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015; 350: aac4383.
Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol 2015; 16: 505–516.
Harrington EA, Fanidi A, Evan GI . Oncogenes and cell death. Curr Opin Genet Dev 1994; 4: 120–129.
Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT . Role of Epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat 2012; 136: 331–345.
Huq MD, Tsai NP, Khan SA, Wei LN . Lysine trimethylation of retinoic acid receptor-alpha: a novel means to regulate receptor function. Mol Cell Proteomics 2007; 6: 677–688.
Zhang Y, Yang X, Gui B, Xie G, Zhang D, Shang Y et al. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J Biol Chem 2011; 286: 42414–42425.
Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y et al. The central role of EED in the orchestration of polycomb group complexes. Nat Commun 2014; 5: 3127.
Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K . Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23: 4061–4071.
Lee TC, Ziff EB . Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J Biol Chem 1999; 274: 595–606.
Rodriguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG et al. Id2 leaves the chromatin of the E2F4-p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J 2006; 398: 431–437.
Wierstra I, Alves J . The c-myc promoter: still MysterY and challenge. Adv Cancer Res 2008; 99: 113–333.
Nicolaidou V, Wong MM, Redpath AN, Ersek A, Baban DF, Williams LM et al. Monocytes induce STAT3 activation in human mesenchymal stem cells to promote osteoblast formation. PLoS ONE 2012; 7: e39871.
Yu H, Jove R . The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 2004; 4: 97–105.
Dasgupta M, Dermawan JK, Willard B, Stark GR . STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA 2015; 112: 3985–3990.
Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA 2010; 107: 21499–21504.
Chung J, Uchida E, Grammer TC, Blenis J . STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol 1997; 17: 6508–6516.
Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006; 366: 2–16.
Tanos B, Rodriguez-Boulan E . The epithelial polarity program: machineries involved and their hijacking by cancer. Oncogene 2008; 27: 6939–6957.
Lock JG, Mamaghani MJ, Shafqat-Abbasi H, Gong X, Tyrcha J, Stromblad S . Plasticity in the macromolecular-scale causal networks of cell migration. PLoS One 2014; 9: e90593.
Nagano M, Hoshino D, Koshikawa N, Akizawa T, Seiki M . Turnover of focal adhesions and cancer cell migration. Int J Cell Biol 2012; 2012: 310616.
Jain N, Iyer KV, Kumar A, Shivashankar GV . Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci USA 2013; 110: 11349–11354.
Vergani L, Grattarola M, Nicolini C . Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int J Biochem Cell Biol 2004; 36: 1447–1461.
Hsieh FC, Cheng G, Lin J . Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun 2005; 335: 292–299.
Gialeli C, Theocharis AD, Karamanos NK . Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 2011; 278: 16–27.
Rangel LB, Agarwal R, D'Souza T, Pizer ES, Alo PL, Lancaster WD et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res 2003; 9: 2567–2575.
Matsuguchi T, Inhorn RC, Carlesso N, Xu G, Druker B, Griffin JD . Tyrosine phosphorylation of p95Vav in myeloid cells is regulated by GM-CSF, IL-3 and steel factor and is constitutively increased by p210BCR/ABL. EMBO J 1995; 14: 257–265.
Natsume A, Ito M, Katsushima K, Ohka F, Hatanaka A, Shinjo K et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res 2013; 73: 4559–4570.
Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007; 21: 1050–1063.
McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108–112.
Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 2012; 8: 890–896.
Chen YT, Zhu F, Lin WR, Ying RB, Yang YP, Zeng LH . The novel EZH2 inhibitor, GSK126, suppresses cell migration and angiogenesis via down-regulating VEGF-A. Cancer Chemother Pharmacol 2016; 77: 757–765.
Kim KH, Roberts CW . Targeting EZH2 in cancer. Nat Med 2016; 22: 128–134.
Toh TB, Lim JJ, Chow EKH . Epigenetics in cancer stem cells. Mol Cancer 2017; 16: 29.
Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res 2007; 67: 10657–10663.
Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 2003; 4: 124–131.
Chen YH, Hung MC, Li LY . EZH2: a pivotal regulator in controlling cell differentiation. Am J Transl Res 2012; 4: 364–375.
Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V . Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 2009; 36: 61–74.
Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 2006; 107: 2170–2179.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006; 125: 301–313.
de Vries NA, Hulsman D, Akhtar W, de Jong J, Miles DC, Blom M et al. Prolonged Ezh2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Rep 2015; 10: 383–397.
Wassef M, Rodilla V, Teissandier A, Zeitouni B, Gruel N, Sadacca B et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev 2015; 29: 2547–2562.
Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao L et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol 2016; 17: 95–103.
De Craene B, Berx G . Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110.
Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008; 28: 4772–4781.
Sauvageau M, Sauvageau G . Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010; 7: 299–313.
Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol 2004; 6: 154–161.
Acknowledgements
We thank S Strömblad for critical reading, discussions and comments on the manuscript; Y Zhang (Harvard University) for the EZH2, SUZ12 and EED baculovirus expression vectors; we thank Y Or for providing BALB/c mice and technical support, LY Lu for advice on statistical analysis and A Sullivan from Obrizus Communications for editing the manuscript. This work was supported by the Singapore Agency for Science Technology and Research (A*STAR) through a Biomedical Research Council (BMRC) grant (10/1/22/19/666) to LN, Singapore Ministry of Education (MOE2009-T2-1-034 and MOE2013-T2-2-038) and Ministry of Health, National Medical Research Council (NMRC-CBRG/0057/201) to I-hS.
Author contributions
NV identified EZH2-mediated Talin methylation sites, initiated the project and established essential collaborations; NV and JFW designed and conducted most of the experiments, including the in vitro methyltransferase assays, identification of the EZH2–VAV interaction mutant and performed experiments in mammary epithelial cells, breast cancer animal models and were also involved in the data interpretation and manuscript preparation; KPT and MSM did computational modeling and predicted residues critical for EZH2–VAV interaction; HHC and VCLL were involved in experiments using breast cancer animal models; EC and JG performed differential expression analysis of RNA-seq data; YHY and SGS performed the SPR analysis; AA and LN produced recombinant octamers and I-hS designed and interpreted the experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Accession codes
ArrayExpress website: http://www.ebi.ac.uk/arrayexpress/help/how_to_search.html#Login
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Venkatesan, N., Wong, J., Tan, K. et al. EZH2 promotes neoplastic transformation through VAV interaction-dependent extranuclear mechanisms. Oncogene 37, 461–477 (2018). https://doi.org/10.1038/onc.2017.309
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.309
This article is cited by
-
Phosphorylation of EZH2 differs HER2-positive breast cancer invasiveness in a site-specific manner
BMC Cancer (2023)
-
AKAP12 promotes cancer stem cell-like phenotypes and activates STAT3 in colorectal cancer
Clinical and Translational Oncology (2023)
-
Inhibition of cytoplasmic EZH2 induces antitumor activity through stabilization of the DLC1 tumor suppressor protein
Nature Communications (2021)
-
Inhibition of EZH2 induces NK cell-mediated differentiation and death in muscle-invasive bladder cancer
Cell Death & Differentiation (2019)