Abstract
Dysregulation of the NF-κB transcription factor occurs in many cancer types. Krüppel-like family of transcription factors (KLFs) regulate the expression of genes involved in cell proliferation, differentiation and survival. Here, we report a new mechanism of NF-κB activation in glioblastoma through depletion of the KLF6 tumor suppressor. We show that KLF6 transactivates multiple genes negatively controlling the NF-κB pathway and consequently reduces NF-κB nuclear localization and downregulates NF-κB targets. Reconstitution of KLF6 attenuates their malignant phenotype and induces neural-like differentiation and senescence, consistent with NF-κB pathway inhibition. KLF6 is heterozygously deleted in 74.5% of the analyzed glioblastomas and predicts unfavorable patient prognosis suggesting that haploinsufficiency is a clinically relevant means of evading KLF6-dependent regulation of NF-κB. Together, our study identifies a new mechanism by which KLF6 regulates NF-κB signaling, and how this mechanism is circumvented in glioblastoma through KLF6 loss.
Similar content being viewed by others
Introduction
The NF-κB transcription factor family is oncogenic through suppression of programmed cell death, and promotion of tumor growth and invasion.1 In tumors, NF-κB can be activated by mutations in its own genes or in its regulating genes.2 In the canonical pathway, NFKBIA (IκBα)3 interacts and sequesters the p65/p50 NF-κB heterodimer in the cytoplasm. Upon various stimuli, NFKBIA is phosphorylated and degraded allowing translocation of NF-κB in the nucleus and transcriptional activation of NF-κB targets. Although both subunits can bind to the DNA, only p65 contains a transcriptional activation domain.4
Mutations and enrichment of specific single-nucleotide polymorphisms and haplotypes of NFKBIA in human cancer suggest a role as tumor suppressor.5, 6, 7, 8 Other genes negatively regulate NF-κB activation, such as the TNF α-induced protein 3 (TNFAIP3; A20), a ubiquitin-editing enzyme which downregulates NF-κB signaling when binding TNFAIP3-interacting proteins 1 and 2 (TNIP1 and TNIP2, respectively).9
We previously found that monoallelic deletion of NFKBIA occurs in about 25% of glioblastomas and convey a dismal clinical prognosis.8 However, aberrant constitutive activation of NF-κB occurs in most glioblastomas,10 suggesting additional mechanisms of NF-κB activation.
KLFs regulate expression of genes involved in signal transduction, proliferation, differentiation, cell death and oncogenesis. KLF6 is a putative tumor suppressor in prostate, colorectal, hepatocellular carcinomas and glioblastoma.11, 12, 13, 14, 15, 16, 17 Deletion of the chromosome region containing KLF6 (10p15) has been reported in glioblastoma,16 whereas mutation analyses of the KLF6 coding region have been controversial.16, 18, 19, 20, 21, 22 KLF6 has been proposed to perform its tumor suppression function by promoting G1 cell cycle arrest mainly through cyclin-dependent kinase inhibitor 1A (CDKN1A) promoter transactivation.15
The KLF6 splice variant sv1 is aberrantly expressed in prostate, ovarian cancer and glioblastoma.16, 23 Upon splicing, KLF6-sv1 lacks a nuclear localization signal; therefore, it cannot transactivate KLF6 targets and supposedly is non-functional.24 Nevertheless, KLF6-sv1 has been shown to promote tumor progression and metastasis in various cancers.25, 26
Here, we employ genome-wide scanning for transcripts co-expressed with NFKBIA, TNFAIP3, TNIP1 and TNIP2 to identify KLF6 as a common transactivator of NF-κB-negative regulatory genes. We demonstrate that KLF6 is frequently inactivated in glioblastoma and propose KLF6 deletion as a new mechanism underlying NF-κB signaling increase in this tumor type.
Results
NF-κB-negative regulators are co-regulated in glioblastoma
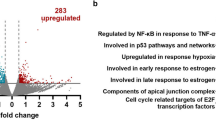
To determine whether deregulation of negative regulators of NF-κB has a role in constitutive NF-κB activation in glioblastoma, we analyzed expressions of the NF-κB regulators NFKBIA, TNFAIP3, TNIP1 and TNIP2 in glioblastoma patients from The Cancer Genome Atlas (TCGA). All regulators showed co-expression, suggesting a common regulation (Figure 1a). We excluded genomic co-mapping (NFKBIA: 14q13.2, TNFAIP3: 6q23.3, TNIP1: 5q33.1 and TNIP2: 4p16.3) of these genes and concurrent gene copy number variation as a cause of their co-expression (Supplementary Figure 1A). To identify common transcriptional regulators, we performed a genome-wide scan for transcripts co-expressed with the four NF-κB-negative regulators. Filtering for cancer-associated transcription factors,27 we identified 18 genes (Figure 1b); 5 of which encoded Rel-like domain-containing proteins forming homo- or heterodimeric NF-κB complexes and inducing NF-κB regulator genes through negative feedback regulation.28, 29 Another seven transcription factors were co-expressed with all the four NF-κB-negative regulators (Figure 1b), suggesting a role in their transcriptional regulation. In a complementary approach, we looked for common transcription factors binding sites by performing in silico promoter binding analyses (MatInspector, Genomatix, Munich, Germany) for all NF-κB-negative regulators and identified 43 transcription factors with binding sites present in all promoters (Supplementary Figure 2).
Negative regulators of NF-κB are co-regulated in glioblastoma. (a) Scatter plot matrix for messenger RNA expression of NF-κB control genes NFKBIA, TNFAIP3, TNIP1 and TNIP2 representing pairwise associations between each of these variables in 188 glioblastomas. Locally weighted least squares smooth fits indicate the appropriateness of the linear regression analyses. The corresponding P-values indicate the statistical significance of these relationships according to estimated slope of the regression line. (false discovery rate adjusted q<1 × 10−5). (b) Genome-wide correlation analysis of the four NF-κB control genes (TNIP1, TNIP2, NFKBIA, TNFAIP3) with other gene transcripts in 413 glioblastomas (significance level: P<1 × 10−5). Significant associations between the NF-κB-negative regulators and Rel-like domain-containing proteins (NFKB1, NFKB2, RELA, c-REL and RELB) are indicated by green edges. Edges connecting the control genes to KLF6 and BCL6, red and blue, respectively. (c) Schematic diagram of predicted binding sites of transcription factors KLF6, BCL6 and NF-κB in the promoter regions of the four NF-κB control genes (NFKBIA, TNFAIP3, TNIP1 and TNIP2).
KLF6 is a clinically relevant putative tumor suppressor
Three transcription factors, B-cell CLL/lymphoma 6 (BCL6), KLF6 and the NF-κB family, were identified in both analyses (Figure 1c). Given the expected feedback between NF-κB and NF-κB control genes, we focused on BCL6, an oncogenic repressor30 and KLF6, a transcriptional activator and tumor suppressor.11 Copy number analysis for KLF6 (10p15) and BCL6 (3q27) revealed heterozygous deletions of KLF6 in 74.5% of tumors, but homozygous deletions in only 0.4% (Figure 2a). BCL6 showed low-level amplification in 7.3% and high-level amplification in 0.7% of the tumors (data not shown). We then assessed KLF6 and BCL6 associations with survival in 406 patients with glioblastoma. We found no survival association for BCL6 amplifications (log-rank P>0.05; data not shown), but patients with KLF6 deletions had significantly shorter progression-free survival and overall survival than those without KLF6 deletions (Figures 2b and c).
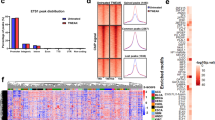
KLF6 is a clinically relevant putative tumor suppressor. (a) Heatmap displaying gene copy number variation analysis for KLF6 (maps to 10p15) in 537 TCGA glioblastomas by circular binary segmentation 36 and Genomic Identification of Significant Targets in Cancer (GISTIC2). The association with four major subtypes (classical, mesenchymal, neural and proneural) of glioblastoma is shown. (b) Kaplan–Meier estimates of overall survival for 406 glioblastoma patients, with patients stratified into two subgroups based on whether their tumor harbored a deletion of KLF6. (c) Kaplan–Meier estimates of progression-free survival in the same patients stratified in the same manner according to KLF6 gene status. (d) Immunoblotting for KLF6 in normal brain tissues, and patient-derived glioblastoma cells. (e) Immunoblotting for KLF6 in normal brain tissues, glioblastoma-derived xenolines. (f) Gene copy number analysis for KLF6 in neural progenitor cells, normal brain tissues and glioblastoma-derived xenolines. (g) Schematic representation of the KLF6 exon structure showing the four coding sequence exons, localization of the nuclear localization signal (NLS) in exon 2 and the new identified mutation (391 G/A, V to M).
Consistent with the high KLF6 heterozygous deletion frequency observed in our analysis, KLF6 expression in glioblastoma was reduced compared with normal brain samples (non-malignant cortical samples obtained from epilepsy surgery, NB) (Figure 2d). In a panel of glioblastoma-derived xenolines characterized for molecular subtype, KLF6 protein did not show association with any specific GBM subtype31 (Figure 2e). Consistently, KLF6 deletion was proportionally distributed among the four subtypes (Figure 2a). Copy number analysis of the same cell panel showed frequent loss of one allele (Figure 2f). However, three xenolines (JX12, JX12T and JX59T) without KLF6 deletion still lacked KLF6 expression hinting at additional silencing mechanisms.
Given previous contradictory results regarding KLF6 mutations in glioblastoma, we sequenced the KLF6 sequence upstream of the ATG (431 bp), CDS exons 1–4 and intron–exon boundaries in 45 glioblastoma samples. We identified several previously reported single-nucleotide polymorphisms (data not shown) and a new mutation (391 G/A, V to M) in one patient (frequency: 0.02) (Figure 2g), confirming that KLF6 is rarely mutated in glioblastoma.22
KLF6 transactivates NF-κB control genes in glioblastoma
To confirm that KLF6 is a transcriptional activator of the four NF-κB-negative regulators, LN229 glioblastoma cells and two brain tumor-derived stem-like cells (BTSCs) (BTSC23, BTSC233) were transduced with a lentivirus expressing empty vector (EV), KLF6-wt or the non-functional KLF6 variant sv1 as a negative control24 (Figures 3a–c and Supplementary Figures 3A–B). We confirmed the lack of nuclear localization by KLF6-sv1, as previously reported24 (Supplementary Figure 3C). RT–PCR confirmed that KLF6-wt, but not KLF6-sv1, induced expression of NFKBIA and TNFAIP3, and to a less extent, of TNIP1 and TNIP2, in all the cell lines tested (Figures 3d–f). Correspondingly, increased NFKBIA and TNFAIP3 protein expression was detected by immunoblotting (Figure 3g), suggesting that NFKBIA and TNFAIP3 are the strongest NF-κB-negative regulators induced by KLF6. Chromatin immunoprecipitation in LN229 cells showed binding of KLF6-wt to all tested promoters, indicating that KLF6-wt can directly regulate the expression of the four NF-κB-negative regulators (Figures 3h and i).
KLF6 induces NF-κB-negative regulators in glioblastoma. (a–c) Infection of LN229 (a), BTSC23 (b) and BTSC233 (c) cells with pCHMWS lentivirus carrying FLAG-tagged KLF6-wt or KLF6-sv1 cDNA, or empty vector (EV), captured by immunoblotting. (d– f) qRT-PCR analysis of relative transcript expression of TNIP1, TNIP2, NFKBIA and TNFAIP3 in LN229 (d), BTSC23 (e), BTSC233 (f) cells expressing EV, KLF6-wt or KLF6-sv1. (g) Immunoblotting of KLF6, TNFAIP3, NFKBIA in LN229 cells expressing EV, KLF6-wt or KLF6-sv1. (h) Schematic representation and position of KLF6 binding sites in the promoters of TNIP1, TNIP2, NFKBIA and TNFAIP3 genes. Stars indicate binding sites validated by chromatin immunoprecipitation. (i) qRT-PCR analysis showing enriched binding of KLF6 to specific promoters in LN229 cells expressing EV, KLF6-wt or KLF6-sv1. The SERPINA1 and OLR1 gene promoters were positive and negative controls, respectively. Relative gene expressions are normalized to EV, representative qRT-PCR of three independent experiments. (d– f). Error bars represent mean±s.d.
Next, we measured the effect of KLF6 on NF-κB pathway by looking at the activity of the canonical NF-κB subunits p50 and p65. In LN229 and BTSC233, KLF6-wt-mediated transcriptional activation of the NF-κB control genes was associated with reduced nuclear binding of the NF-κB subunits to a κB-responsive sequence (Figures 4a and b and Supplementary Figure 4A). Moreover, KLF6-wt, but not KLF6-sv1, reduced the expression of a NF-κB reporter construct (Figure 4c). Nuclear localization of p65 in LN229 and BTSC23 cells was reduced as a consequence of KLF6-wt expression as shown by immunoblotting of cytoplasmic and nuclear cell extracts (Figures 4d and e), and p65 immunostaining in LN229 cells (Figure 4f). Then, we measured the expression of a panel of NF-κB target genes previously implicated in glioblastoma by RT–PCR (Figure 4g and Supplementary Figures 4B and C).32, 33, 34, 35, 36, 37, 38, 39 In all tested cells, the majority of the genes were downregulated upon KLF6-wt overexpression, whereas KLF6-sv1 had no effect. Downregulation of NF-κB targets MMP9, OLIG2 and YKL40 was confirmed by western blot (Figure 4h). Finally, to demonstrate that KLF6-induced downregulation of NF-κB targets was due to upregulation of NF-κB-negative regulators, we silenced NFKBIA in LN229 cells previously transduced with EV, KLF6-wt or KLF6-sv1 (Figures 4i and j and Supplementary Figure 4D). As shown in Figure 4j, NFKBIA knockdown completely or partially rescued KLF6-wt effect on the majority of the tested genes; furthermore, although not always statistically significant, it seemed to prevent KLF6-mediated upregulation of the other NF-κB-negative regulators (TNIP1, TNIP2 and TNFAIP3, Supplementary Figure 4E), suggesting that NF-κB could mediate a positive feedback loop by downregulating its negative regulators as previously described.40, 41 Altogether, these results further confirm a role of KLF6 in NF-κB activation through transactivation of NF-κB control genes.
KLF6 inhibits NF-κB signaling in glioblastoma. (a–b) Analysis of NF-κB subunit p50 (a) and p65 (b) activation in nuclear and cytoplasmic extracts of LN229 cells expressing empty vector EV, KLF6-wt or KLF6-sv1. Data are normalized to protein concentration and are expressed as mean±s.d. (c) Luciferase assay of NF-κB subunit p65 in TNFα-stimulated LN229 cells expressing EV, KLF6-wt or KLF6-sv1. (d–e) Immunoblotting analysis for NF-κB p65 and KLF6 in nuclear and cytoplasmic extracts of TNFα-stimulated LN229 (d) and BTSC23 (e) cells expressing EV, KLF6-wt or KLF6-sv1. (f) Immunofluorescence staining of NF-κB subunit p65 in TNFα-stimulated LN229 cells expressing EV, KLF6-wt or KLF6-sv1. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). The right panel shows the corresponding quantification of nuclear:cytoplasmic ratio. The scale bar represents 30 μm (low mag) and 10 μm (high mag). (g) qRT-PCR analysis of relative transcript expression of NF-κB targets in LN229 cells. Relative gene expressions are normalized to EV, representative qRT-PCR of three independent experiments is shown. Error bars represent mean±s.d. (n=3 qPCR replicates) (j). (h) Immunoblotting of MMP9, OLIG2, YKL40 in LN229 and BTSC23 cells expressing EV, KLF6-wt or KLF6-sv1. *Represents an aspecific band. (i) Immunoblotting of NFKBIA and KLF6 in LN229 cells transduced as indicated. (j) qRT-PCR analysis of relative transcript expression of NF-κB targets in LN229 cells transduced as indicated. Error bars represent mean±s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.005.
KLF6 induces the expression of neural-like genes and inhibits the malignant phenotype of glioblastoma in vitro
LN229 cells expressing KLF6-wt had an elongated cell body reminiscent of neural cells (Figure 5a). These morphological changes were accompanied by upregulation of neuronal marker β3-tubulin (TUBB3) and neurofilament medium polypeptide (NEFM); by contrast, expression of glial marker glial fibrillary acidic protein (GFAP) and the stem cell marker NESTIN was not affected (Figures 5b–d). Immunoblotting and immunofluorescence confirmed high expression of TUBB3 upon KLF6-wt expression (Figures 5c and d). Moreover NESTIN was reduced while no changes in GFAP and two synaptic markers, PSD95 and SYP (Synaptophysin), were observed by immunostaining (Supplementary Figure 5A). These data suggests that KLF6-wt overexpression in LN229 activates the expression of some neuronal markers (TUBB3 and NEFM) although the differentiation appeared uncomplete due to lack of synaptic markers.
Wild-type KLF6 induces neural-like differentiation in LN229 cells. (a) Microphotographs of green fluorescent protein-positive LN229 cells expressing empty vector (EV), KLF6-wt or KLF6-sv1 after lentiviral infection. (b) qRT-PCR analysis of neuronal (TUBB3 and NEFM), astrocytic (GFAP) and stem cell (NESTIN) markers in LN229 cells expressing EV, KLF6-wt or KLF6-sv1. (c) Immunoblotting of TUBB3 in LN229 cells expressing EV, KLF6-wt or KLF6-sv1. (d) Corresponding immunofluorescence staining for neuronal marker TUBB3 in LN229 cells expressing EV, KLF6-wt or KLF6-sv1. Nuclei were stained with DAPI. The scale bar represents 100 μm (a) or 50 μm (d). Relative gene expressions are normalized to EV, a representative qRT-PCR of three independent experiments is shown (b). Error bars represent mean±s.d.
KLF6-wt overexpression in two BTSCs (BTSC23, BTSC233) led to morphological changes similar to LN229 (Figure 6a and Supplementary Figure 6A). RT–PCR analyses in BTSC23 cells showed increased TUBB3 and NEFM expression, whereas GFAP expression was slightly reduced and NESTIN expression was unaffected (Figure 6b). Similar results for TUBB3 and GFAP were obtained in BTSC233 cells (Supplementary Figure 6B), whereas NEFM was expressed at very low levels in all the samples (data not shown). TUBB3 and NEFM increase in BTSC23 was confirmed at the protein level (Figures 6c–e, Supplementary Figures 6C and D). Interestingly, in BTSC233, but not in BTSC23, cells higher levels of GFAP were detected by immunostaining (Supplementary Figure 5B and Supplementary Figure 6E), suggesting that glial markers could also be regulated in a cell-dependent context. PSD95 and SYP were also upregulated upon KLF6-wt expression in BTSC233 and BTSC23 cells, respectively (Supplementary Figure 5B and Supplementary Figures 6F and G). Overall, these data indicate that, although the expression of specific neural markers can differ in various glioblastoma cells, KLF6-wt and ensuing NF-κB inhibition appear to induce a similar pattern of expression of neural genes and loss of stem marker NESTIN, suggesting the activation of an aberrant differentiation program.
Wild-type KLF6 induces neural-like differentiation in BTSC23 cells. (a) Microphotographs of GFP-positive BTSC23 cells expressing empty vector (EV), KLF6-wt or KLF6-sv1, after lentiviral infection. (b) qRT-PCR analysis of neuronal (TUBB3, NEFM), astrocytic (GFAP) and stem cell (NESTIN) markers in BTSC23 cells expressing EV, KLF6-wt or KLF6-sv1. (c) Immunoblotting of TUBB3 in BTSC23 cells expressing EV, KLF6-wt or KLF6-sv1. (d) Immunofluorescence staining and relative quantification for TUBB3 in BTSC23 cells expressing EV, KLF6-wt or KLF6-sv1. Nuclei were stained with DAPI. (e) Immunofluorescence staining using Tyramide Signal Amplification and relative quantification for NEFM in BTSC23 cells expressing EV, KLF6-wt or KLF6-sv1. Nuclei were stained with DAPI. *P<0.05, ***P<0.001. The scale bar represents 50 μm (a, e) or 100 μm (d). Relative gene expressions are normalized to EV, a representative qRT-PCR of three independent experiments is shown (b). Error bars represent mean±s.d.; n=10 in d–e.
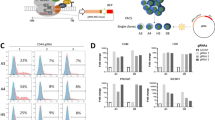
Prolonged expression of KLF6-wt, but not KLF6-sv1, induced a senescent-like phenotype in both LN229 and BTSC23 cells, highlighted by β-galactosidase staining (Figures 7a–f). Cell cycle analysis revealed accumulation of cells in phase G1–G0 and concomitant reduction of cells in phases S and G2/M, upon KLF6-wt expression (Supplementary Figure 7A). In contrast, KLF6-sv1 overexpression prolonged S-phase (Supplementary Figure 7A). In BTSC23 cells, KLF6-wt overexpression led to accumulation of cells in G2/M (Supplementary Figure 7B). Consistent with the G1 arrest observed in LN229 cells, KLF6-wt overexpression led to upregulation of CDKN1A expression in LN229 and BTSC23 cells (Supplementary Figures 7C–D). In accordance with a reduced number of cells in S-phase, KLF6-wt reduced cell growth in both LN229 and BTSC23 cells (Figures 7g and h). KLF6-wt cells appeared less viable compared with control or KLF6-sv1-expressing cells (Supplementary Figures 7E and F) suggesting that KLF6-wt could cause cell death. Only a small increase in apoptosis was observed in BTSC233 but not in LN229 cells upon KLF6-wt overexpression (Figures 7i and j). Interestingly, in BTSC233 cells, KLF6-sv1 reduced apoptosis supporting previous studies that proposed KLF6-sv1 as an oncogenic variant, inhibiting pro-apoptotic factors.25, 42, 43 Finally, KLF6-wt but not KLF6-sv1 overexpression reduced cell migration and invasion in LN229 and BTSC233 cells (Figures 7k–o and Supplementary Figure 7G). Interestingly, LN229 cells transduced with KLF6-sv1 were more proliferative and invasive compared with control cells (Figures 7a and o), supporting KLF6-sv1 oncogenic role.
KLF6 inhibits the malignant phenotype of glioblastoma in vitro. (a– f) Cellular senescence in LN229 (a), BTSC23 (c), BTSC233 (e) cells expressing empty vector (EV), KLF6-wt or KLF6-sv1 assessed by senescence-associated β-galactosidase detection and corresponding quantification of stained cells in LN229 (b), BTSC23 (d), BTSC233 (f). (g–h) Quantification of EdU staining in LN229 (g) and BTSC233 (h) cells expressing EV, KLF6-wt or KLF6-sv1. (i–j) Analysis of cell death apoptotic activity in LN229 (i) and BTSC233 (j) cells. Readings were normalized to blank and are expressed as mean±s.d. (k–n) Scratch assay in LN229 (k) and BTSC233 (m) cells expressing EV, KLF6-wt or KLF6-sv1, and corresponding quantification in LN229 (l) and BTSC233 (n) cells. (o) Quantification of invading LN229 cells expressing EV, KLF6-wt or KLF6-sv1 in a matrigel invasion assay. *P<0.05, **P<0.01, ***P<0.001. Scale bars represent 100 μm (a, c, e), 200 μm (k) or 50 μm (m). Error bars represent mean±s.d. (n=3).
Wild-type KLF6 delays gliomagenesis in vivo
Next, we examined whether KLF6-wt and KLF6-sv1 affected tumor formation in vivo. Ectopic expression of KLF6-wt attenuated tumor growth as evidenced by reduced bioluminescence imaging (BLI analysis) 3 weeks after cell injection (P=0.001, t-test) (Figure 8a). Moreover, KLF6-wt overexpression improved survival (Figure 8b), although tumors eventually developed in all the three groups and appeared similar in size (Supplementary Figure 8A). Analysis of the proliferation marker Ki67 (MIB1) did not reveal difference in proliferation, suggesting that perhaps, at the experimental end point the more proliferative cells have been selected and enriched (Supplementary Figure 8B). Supporting our hypothesis that KLF6 regulates NFKBIA expression, NFKBIA was moderately upregulated in KLF6-wt tumors (Supplementary Figure 8C). As observed in vitro, KLF6-wt tumors expressed lower levels of NESTIN and appeared more differentiated as indicated by higher levels of TUBB3 and GFAP (Figures 8c–e). In addition, MMP9 was reduced consistently with our previous observation (Figure 8f). Our in vivo results point toward KLF6 as a tumor suppressor in glioblastoma inhibiting NF-κB activity, causing loss of stemness and promoting differentiation.
KLF6 delays gliomagenesis in vivo. (a) Representative in vivo BLI analysis in NOD/SCID mice 3 weeks after intracranial injection of LN229 cells infected with lentiviral vector carrying KLF6-wt cDNA, KLF6-sv1 cDNA or EV, and pCHMWS-fLuc to express a luciferase gene for BLI detection. (b) Kaplan–Meier estimates of survival in animal subgroups injected as described in a (10 animal per group). (c– f) Immunofluorescence staining and relative quantification for TUBB3 (c), GFAP (d), NESTIN (e), MMP9 (f), in tumors resulting from intracranial injection of LN229 cells injected as described in a. Nuclei were stained with DAPI. *P<0.05, **P<0.01, ***P<0.001. Scale bars represent 100 μm (c–f). Error bars represent mean±s.d.; n=9 in b, n=10 in c–f.
Discussion
In this study we have identified a previously unappreciated NF-κB regulatory pathway that involves transactivation of NF-κB-negative regulatory genes by the tumor suppressor KLF6 in glioblastoma. These findings reinforce prior evidence of a role of the NF-κB pathway in glioma progression.38, 44, 45 Distinct from others, our proposed mechanism of NF-κB deregulation does not involve mutations of NF-κB pathway components.2
Our data support prior studies indicating that KLF6 is a tumor suppressor in human cancer11, 42 and provide a mechanism for its function in glioblastoma. Moreover, in agreement with a previous study in hepatocellular carcinoma,13 we found that loss of one KLF6 allele significantly shortens both progression-free and overall patient survival. We also observed that the remaining KLF6 allele is rarely mutated in glioblastoma, as previously reported.22 Altogether, these data suggest that KLF6 exhibits haploinsufficiency in glioblastoma. However, as all the samples analyzed lacked KLF6 expression, it is possible that other mechanisms are involved in suppressing KLF6 expression. In accordance with this hypothesis, KLF6 has been recently shown to be silenced by increased trimethylation of histone H3 at Lys9 (H3K9me3) levels in dedifferentiated liposarcomas.46 As such, further studies would be required to determine whether additional epigenetic mechanisms silence KLF6 in glioblastoma and/or whether they are alternative to genomic alterations.
Co-activator roles of KLF6 in inducing NF-κB targets were recently proposed.47, 48 Although these data diverge from our proposed model, it is possible that KLF6 might function as NF-κB activator or inhibitor in different tissues and/or in response to different stimuli. As the majority of our experiments were performed in absence of stimulation, it is possible that NF-κB activation by other factors might counteract KLF6-mediated effect. More studies would be necessary to further investigate the relationship between KLF6 and NF-κB in different cell models and condition.
Consistent with previous data,15 we show that KLF6 induces CDKN1A expression in LN229 cells and concomitantly accumulates cells in the G1 phase. In BTSC23 cells, KLF6-wt caused accumulation of cells in G2 phase, which could reflect the different nature of this tumor cell type. This is consistent with previous data in colon cancer cell lines showing that the number of cells arresting in G1 or G2 can be cell type dependent due to regulation by different checkpoint mechanisms.49 As CDKN1A inhibits both G1 and G2 cell cycle phase progression by binding to and inhibiting CDK2 and CDK1,50 our observation that KLF6-wt mediates G2 arrest in BTSC23 cells is also consistent with the induction of CDKN1A by KLF6. Hoeferlin et al.49 suggest that the lower level of CDKN1A expression in HCT116 could be insufficient to cause G1 arrest. Interestingly, we also observed a lower induction of CDKN1A in BTSC23, which accumulated in G2 upon KLF6-wt overexpression. However, it is also possible that CDKN1A-independent mechanisms leading to accumulation of cells in G2 might be activated in response to KLF6-wt.
We observed decreased cell viability in LN229-KLF6-wt cells, suggesting that KLF6 might induce cell death; however, only a small increase in apoptosis was observed upon KLF6 overexpression in BTSC233 cells, which suggests the involvement of additional cell death mechanisms such as mitotic catastrophe, which has been associated with cell senescence.51
We show that KLF6 overexpression often reduced NESTIN and increased neural genes expression in glioblastoma cells and LN229-KLF6-wt derived tumors, suggesting that KLF6-wt could reduce stemness and induce neural differentiation. The enhanced ability of KLF6-wt expressing cells to activate an aberrant/incomplete neural-like differentiation program is in line with a previously reported role of NF-κB in mesenchymal differentiation of glioblastoma-initiating cells.52 Interestingly, blockade of NF-κB in these cells is also associated with increased senescence, similarly to what we observed in our study.52 One could argue that senescence occurs as a consequence of activating an aberrant KLF6-mediated neural differentiation pathway in glioblastoma cells growing under proliferative conditions. Higher levels of GFAP protein, but not RNA, were observed upon KLF6 overexpression in BTSC233 cells. As this increase was not accompanied by upregulation of the GFAP transcript (Supplementary Figure 5B), it is possible that KLF6 overexpression might cause GFAP posttranslational modifications affecting its stability. Interestingly, GFAP phosphorylation has been shown to regulate its stability.53 More studies would be necessary to investigate the role of KLF6 in GFAP protein regulation. We show that KLF6 regulated NF-κB via NF-κB regulators requires KLF6 localization into the nucleus and binding to their promoters. In agreement, the splice variant sv1 lacking the nuclear localization domain does not affect NF-κB activation. Interestingly, overexpression of KLF6-sv1 leads to a significant increase in the percent of cells in S-phase, cellular proliferation and cell invasion, consistent with the proposed role of KLF6-sv1 as an oncogenic variant, which drives metastasis and is associated with poor survival in prostate cancer.25 Our data suggest that KLF6-sv1-mediated oncogenic functions do not require translocation into the nucleus and repression of NF-κB-negative regulators. Further studies would be required to characterize the mechanism of KLF6-sv1 function in tumorigenesis and metastasis.
Collectively, our data are the first to identify KLF6 as a negative regulator of NF-κB and indicate that disruption of this system can occur by loss of function of KLF6 through haploinsufficiency. Our findings could potentially translate the use of NF-κB inhibitors in combination to drugs that allow KLF6 re-expression. Interestingly, it was shown that targeting the KLF6 regulator FOXO1 and EGFR with trifluoperazine hydrochloride, a FOXO1 nuclear export inhibitor, and erlotinib, a small-molecule inhibitor of EGFR signaling, leads to KLF6 re-expression.25 In the future, it would be interesting to test the effect of these drugs, alone or in combination with NF-κB inhibitors, on glioblastoma cell proliferation and invasion, in vitro and in vivo.
Materials and methods
Tumor samples and patients
Samples were collected at the University of Freiburg and the University of Alabama at Birmingham (UAB), under institutional review board (IRB)-approved guidelines. Written informed consent was obtained from all patients. About 537 glioblastoma samples from TCGA (http://cancergenome.nih.gov/) were used as discovery set. Differences in sample sizes for the different analyses reflect the availability of patient data for the different inferred molecular levels (messenger RNA, copy number and so on), platforms and incomplete overlap between these data sets. Raw Affymetrix Genome-Wide Human SNP Array 6.0 and Agilent Human Genome CGH Microarray 244A gene dosage data, Affymetrix Human Genome U133 Plus 2.0 Array and Agilent 244K Custom Gene Expression data, and Affymetrix Human Exon 1.0 ST Array exon-specific expression data, and clinical data were retrieved from the TCGA upon National Human Genome Research Institute (NHGRI) approval and pre-processed for downstream analyses.
Cell lines and cell culture
LN229 glioblastoma cells were obtained from the ATCC and grown according to basic cell culture techniques. Patient-derived glioblastoma stem cells (BTSCs) were prepared from tumor specimens under IRB-approved guidelines and grown as previously described.54 All cells were mycoplasma-free.
Gene copy number variation analysis
Gene-level copy number variation was estimated using the CBS algorithm55 from the 'snapCGH' package for R. Gene dosage segments were classified as chromosomal ‘gain’ or ‘loss’ if the absolute value of the predicted dosage was more than 0.75 times the interquartile range of the difference between observed and predicted values for each region. Copy number variation data processed using the GISTIC2 algorithm56 were retrieved from the Broad Institute at http://gdac.broadinstitute.org/runs/analyses__2012_03_21/data/GBM/20120321/. Genes mapped onto the human genome coordinates using the University of Santa Cruz Biotechnology (Dallas, TX, USA) (UCSC) cgData HUGO probeMap were visualized using the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/).
Quantitative real-time PCR
RNA was prepared as described before.54 Quantitative real-time PCR (qRT-PCR) was performed in triplicate using SYBR Green or pre-validated TaqMan assays (Applied Biosystems, Thermo Fisher, Waltham, MA, USA): KLF6-wt: HKLF6E2E3wt, KLF6-sv1: HS01062724, CHI3L1: HS00609691, CDKN1A: HS00355782. Primer sequences are listed in Supplementary Table 1. Experiments were validated three times, a representative experiment (mean refers to technical replicates) or the average of three experiments was shown.
Immunoblotting and immunofluorescence
Antibodies for immunoblotting and immunostaining are listed in Supplementary Table 5. Secondary labeling of NEFM antibody was performed with Tyramide Signal Amplification Kit (PerkinElmer, Waltham, MA, USA) according to the manufacturer’s instructions. Samples were counterstained with DAPI. Pictures were acquired using an Axiovert Microscope (Zeiss, Oberkochen, Germany) and FSL confocal microscope (Olympus, Tokyo, Japan). Axiovision AXIOVS40 V4.8.0.0 (Carl Zeiss, Oberkochen, Germany) or Fluoview FV10-ASW3.1 (Olympus) software was used for image processing and quantifications.
Vectors and lentiviral infection
Lentiviral infections were performed as previously described54 using the lentiviral vector pCHMWS (kind gift from Veerle Baekelandt, University of Leuven) expressing KLF6-wt, KLF-sv1 or EV. Knockdown of NFKBIA was obtained with a shRNA lentiviral vector (pGIPZ, Open Biosystem Clone ID _V3LHS_410687). Cloning primers are listed in Supplementary Table 3.
Chromatin immunoprecipitation
Promoter analysis was performed with the MatInspector software (www.Genomatix.de). Primers were designed using the Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and are listed in Supplementary Table 2. Chromatin immunoprecipitation was performed three times as previously described57 using KLF6 antibody (R-173, Santa Cruz Biotechnology) or normal rabbit immunoglobulins (Santa Cruz Biotechnology). Eluted DNA was analyzed by absolute qRT-PCR. Amplified product was expressed as a percentage of the input for each condition. The SERPINA1 and OLR1 gene promoters were positive and negative controls, respectively.58
Mutational analysis
KLF6 sequence analysis was carried out on a 3730 DNA Analyzer (Applied Biosystems). Primer sequences are listed in Supplementary Table 4.
Nuclear and cytoplasmic extracts, and NF-κB DNA binding assay
Cytoplasmic and nuclear extracts were prepared using a Nuclear Extract Kit (Active Motif, La Hulpe, Belgium) and processed using the TransAM assay p50 and p65 kits (Activ Motif, La Hulpe, Belgium) according to the manufacturer's instructions.
Luciferase assay
Luciferase analysis was performed using the Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. About 2.5 × 105 cells were seeded in six-well plates prior to transfection with NF-κB-responsive vector (3xkB.luc) or control vector together with renilla luciferase vector. Six independent transfections were done using Lipofectamine according to the manufacturer’s instruction (Invitrogen, Thermo Fisher). Four to six hours after transfection, cells were infected for 72 h and then stimulated with 10 ng/ml recombinant human TNFα (R&D systems, Minneapolis, MN, USA) for 6 h. Luciferase activity was determined using a Thermo Scientific Appliskan luminometer (Termo Fisher). All data were reported relative to luciferase activity (firefly/renilla).
Cell viability assay
Cells were harvested 5 days after lentiviral transduction and 2000 cells per well were seeded in 96-well plates. Proliferation was measured at 24-h intervals up to 6 days (LN229) or at 0, 1, 4, 5, 7, 8 days (BTSC23) by (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)) conversion (Roche, Mannheim, Germany) at 550/700 nm on a plate reader, according to the manufacturer’s instructions. Assays were conducted in six replicates in two independent experiments.
EdU cell proliferation assay
Cell proliferation was assessed using the EdU-Click594 Cell Proliferation Imaging Kit (Baseclick GmbH, Neuried, Germany) according to the manufacturer's instructions. About 2 × 104 cells were seeded on glass coverslips (LN229) or laminin-coated glass coverslips (BTSCs) in a 24-well plate. Pictures were acquired using an Axiovert Microscope (Zeiss). Assays were conducted in triplicates in two independent experiments.
Cell cycle analysis
For flow cytometry analysis cells were infected as described above. At 24-h intervals, floating and attached cells were harvested and fixed with 70% ethanol for 1 h at 4 °C. Fixed cells were treated with 0.2 mg/ml RNase A (Sigma, St Louis, MO, USA) for 1 h at 37 °C, stained with 10 μg/ml propidium iodide (PI; Sigma) and analyzed on a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Percentages of cells in G0–G1, S and G2–M phases were determined (Flow Jo, Ashland, OR, USA). Three independent experiments were performed.
Senescence assay
Senescence was assessed using the Senescence Detection kit (Calbiochem, Darmstadt, Germany) according to the manufacturer’s instructions. Cells were seeded in triplicates, fixed and stained 5 days upon lentiviral transduction. Pictures were acquired using an Axioimager 2 Microscope (Zeiss). β-galactosidase-positive cells were counted and expressed as a percentage of total cells. The assay was validated in two independent experiments.
Caspase 3/7 activity assay
For caspase assay, 2000 cells per well were seeded in 96-well plates (Promega). Transduced and positive control cells (treated with 2.5 μm VP16 (Etoposide) for 24 h) were seeded in triplicates and incubated with Caspase 3/7 R Glo reagent and analyzed in a plate-reading luminometer as per manufacturer’s instruction. The assay was validated in two independent experiments.
Migration and invasion assays
Migration and invasion assays were performed as described before.54 Images of migrating cells were taken every 24 h. For BTSC233 cells, laminin-coated (Invitrogen, Thermo Fisher, 4 μg/ml) 60 mm dishes containing a culture insert (Ibidi, Martinsried, Germany) were used. For invasion assay, 2.5 × 104 cells were seeded in four replicates in the upper compartment. Pictures were acquired using an Axioimager 2 Microscope (Zeiss). The assays were validated in two independent experiments.
Intracranial injection and bioluminescence analysis
Intracranial injections were performed in NOD/SCID mice (Charles River Laboratory) in accordance with the directive 86/609/EEC of the European Parliament, following approval by regional authorities, as described before.59 Mice were randomized in experimental groups by a blinded operator. Sample size in each group (10) was chosen based on one-way analysis of variance to give 80% power to detect significant difference with three groups. Animals were monitored daily until the development of neurological symptoms by a blinded operator. Animals which did not show tumors were excluded from the analysis. Bioluminescence imaging was conducted with a CCD camera (IVIS, Xenogen, Caliper life Science, Maiz, Germany). The data were expressed as photon-flux (photons/sec/cm2/steradian).
Histological analysis and immunohistochemistry
Histology was performed as previously described.60, 54 Sections were incubated with primary antibodies listed in Supplementary Table 5. Nuclei were counterstained with DAPI. Pictures were acquired using a FSL confocal microscope (Olympus) and AxioImager 2 (Zeiss). Fluoview FV10-ASW3.1 (Olympus) and Adobe Photoshop CS5 software (San Jose, CA, USA) were used for image processing and quantifications. Quantification of Ki67 staining was done using IHC profiler in ImageJ.61
Statistical analyses
Linear regression analyses and graphical model validation were executed using R software (Vienna, Austria). Scatterplots and locally weighted least squares smooths were used to confirm the suitability of linear regression analyses, and statistical significance was assessed according to the P-value for the estimated slope of the regression line. Survival curves were estimated by the Kaplan–Meier product-limit method, and qsurvival distributions were compared across the groups using the log-rank test. Univariate and multivariate Cox proportional hazards regression analyses were carried out with progression-free survival or overall survival as the dependent variable and KLF6 gene dosage or KLF6 gene expression as the primary predictor. Two-way contingency table analysis, unpaired t-test and Wilcoxon rank-sum test were used as appropriate. Odds ratios in the two-way contingency table analysis and 95% confidence intervals were computed using Woolf’s method for variance estimation. Comparison of treatment was done by paired two-ways Student’s t-test. P-values <0.05 were considered significant. Error bars represent s.d.
References
Karin M . Nuclear factor-kappaB in cancer development and progression. Nature 2006; 441: 431–436.
Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009; 459: 717–721.
Jacobs MD, Harrison SC . Structure of an IkappaBalpha/NF-kappaB complex. Cell 1998; 95: 749–758.
May MJ, Ghosh S . Rel/NF-kappa B and I kappa B proteins: an overview. Semin Cancer Biol 1997; 8: 63–73.
Cabannes E, Khan G, Aillet F, Jarrett RF, Hay RT . Mutations in the IkBa gene in Hodgkin's disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 1999; 18: 3063–3070.
Jungnickel B, Staratschek-Jox A, Brauninger A, Spieker T, Wolf J, Diehl V et al. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin's lymphoma. J Exp Med 2000; 191: 395–402.
Osborne J, Lake A, Alexander FE, Taylor GM, Jarrett RF . Germline mutations and polymorphisms in the NFKBIA gene in Hodgkin lymphoma. Int J Cancer 2005; 116: 646–651.
Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP et al. NFKBIA deletion in glioblastomas. N Engl J Med 2011; 364: 627–637.
Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol 1999; 145: 1471–1482.
Raychaudhuri B, Han Y, Lu T, Vogelbaum MA . Aberrant constitutive activation of nuclear factor kappaB in glioblastoma multiforme drives invasive phenotype. J Neurooncol 2007; 85: 39–47.
Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001; 294: 2563–2566.
Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology 2004; 126: 1090–1103.
Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology 2004; 40: 1047–1052.
Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O et al. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res 2004; 64: 3838–3843.
Kimmelman AC, Qiao RF, Narla G, Banno A, Lau N, Bos PD et al. Suppression of glioblastoma tumorigenicity by the Kruppel-like transcription factor KLF6. Oncogene 2004; 23: 5077–5083.
Camacho-Vanegas O, Narla G, Teixeira MS, DiFeo A, Misra A, Singh G et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer 2007; 121: 1390–1395.
Yao YL, Ma J, Wang P, Xue YX, Li Z, Zhao LN et al. miR-101 acts as a tumor suppressor by targeting Kruppel-like factor 6 in glioblastoma stem cells. CNS Neurosci Ther 2015; 21: 40–51.
Jeng YM, Hsu HC . KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer 2003; 105: 625–629.
Montanini L, Bissola L, Finocchiaro G . KLF6 is not the major target of chromosome 10p losses in glioblastomas. Int J Cancer 2004; 111: 640–641.
Kohler B, Wolter M, Blaschke B, Reifenberger G . Absence of mutations in the putative tumor suppressor gene KLF6 in glioblastomas and meningiomas. Int J Cancer 2004; 111: 644–645.
Koivisto PA, Zhang X, Sallinen SL, Sallinen P, Helin HJ, Dong JT et al. Absence of KLF6 gene mutations in human astrocytic tumors and cell lines. Int J Cancer 2004; 111: 642–643.
Masica DL, Karchin R . Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res 2011; 71: 4550–4561.
DiFeo A, Martignetti JA, Narla G . The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat 2009; 12: 1–7.
Rodriguez E, Aburjania N, Priedigkeit NM, DiFeo A, Martignetti JA . Nucleo-cytoplasmic localization domains regulate Kruppel-like factor 6 (KLF6) protein stability and tumor suppressor function. PLoS One 2010; 5: e12639.
Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J et al. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest 2008; 118: 2711–2721.
Hatami R, Sieuwerts AM, Izadmehr S, Yao Z, Qiao RF, Papa L et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med 2013; 5: 169ra12.
Sadelain M, Papapetrou EP, Bushman FD . Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer 2012; 12: 51–58.
Lai TY, Wu SD, Tsai MH, Chuang EY, Chuang LL, Hsu LC et al. Transcription of Tnfaip3 is regulated by NF-kappaB and p38 via C/EBPbeta in activated macrophages. PLoS One 2013; 8: e73153.
Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D . The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev 1993; 7: 1266–1276.
Phan RT, Dalla-Favera R . The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004; 432: 635–639.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Tracey L, Perez-Rosado A, Artiga MJ, Camacho FI, Rodriguez A, Martinez N et al. Expression of the NF-kappaB targets BCL2 and BIRC5/Survivin characterizes small B-cell and aggressive B-cell lymphomas, respectively. J Pathol 2005; 206: 123–134.
Angileri FF, Aguennouz M, Conti A, La Torre D, Cardali S, Crupi R et al. Nuclear factor-kappaB activation and differential expression of survivin and Bcl-2 in human grade 2-4 astrocytomas. Cancer 2008; 112: 2258–2266.
Tsunoda K, Kitange G, Anda T, Shabani HK, Kaminogo M, Shibata S et al. Expression of the constitutively activated RelA/NF-kappaB in human astrocytic tumors and the in vitro implication in the regulation of urokinase-type plasminogen activator, migration, and invasion. Brain Tumor Pathol 2005; 22: 79–87.
Watanabe A, Mabuchi T, Satoh E, Furuya K, Zhang L, Maeda S et al. Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: participation of nuclear factor-kappaB in upregulation of syndecan-1 expression. J Neurooncol 2006; 77: 25–32.
Eckhardt I, Roesler S, Fulda S . Identification of DR5 as a critical, NF-kappaB-regulated mediator of Smac-induced apoptosis. Cell Death Dis 2013; 4: e936.
Jennewein C, Karl S, Baumann B, Micheau O, Debatin KM, Fulda S . Identification of a novel pro-apoptotic role of NF-kappaB in the regulation of TRAIL- and CD95-mediated apoptosis of glioblastoma cells. Oncogene 2012; 31: 1468–1474.
Lee DW, Ramakrishnan D, Valenta J, Parney IF, Bayless KJ, Sitcheran R . The NF-kappaB RelB protein is an oncogenic driver of mesenchymal glioma. PLoS One 2013; 8: e57489.
Lee YD, Cui MN, Yoon HH, Kim HY, Oh IH, Lee JH . Down-modulation of Bis reduces the invasive ability of glioma cells induced by TPA, through NF-kappaB mediated activation of MMP-9. BMB Rep 2014; 47: 262–267.
Krikos A, Laherty CD, Dixit VM . Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 1992; 267: 17971–17976.
Gurevich I, Zhang C, Encarnacao PC, Struzynski CP, Livings SE, Aneskievich BJ . PPARgamma and NF-kappaB regulate the gene promoter activity of their shared repressor TNIP1. Biochim Biophys Acta 2012; 1819: 1–15.
DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res 2008; 68: 965–970.
Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G et al. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res 2009; 69: 4733–4741.
Westhoff MA, Zhou S, Nonnenmacher L, Karpel-Massler G, Jennewein C, Schneider M et al. Inhibition of NF-kappaB signaling ablates the invasive phenotype of glioblastoma. Mol Cancer Res 2013; 11: 1611–1623.
Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013; 24: 331–346.
Keung EZ, Akdemir KC, Al Sannaa GA, Garnett J, Lev D, Torres KE et al. Increased H3K9me3 drives dedifferentiated phenotype via KLF6 repression in liposarcoma. J Clin Invest 2015; 125: 2965–2978.
Zhang Y, Lei CQ, Hu YH, Xia T, Li M, Zhong B et al. Kruppel-like factor 6 is a co-activator of NF-kappaB that mediates p65-dependent transcription of selected downstream genes. J Biol Chem 2014; 289: 12876–12885.
Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH . Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem 2014; 289: 10318–10329.
Hoeferlin LA, Oleinik NV, Krupenko NI, Krupenko SA . Activation of p21-dependent G1/G2 arrest in the absence of DNA damage as an antiapoptotic response to metabolic stress. Genes Cancer 2011; 2: 889–899.
Abbas T, Dutta A . p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–414.
Eom YW, Kim MA, Park SS, Goo MJ, Kwon HJ, Sohn S et al. Two distinct modes of cell death induced by doxorubicin: apoptosis and cell death through mitotic catastrophe accompanied by senescence-like phenotype. Oncogene 2005; 24: 4765–4777.
Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Lafarga M, Berciano MT, Aldaz B et al. Blockade of the NFkappaB pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene 2011; 30: 3537–3548.
Takemura M, Gomi H, Colucci-Guyon E, Itohara S . Protective role of phosphorylation in turnover of glial fibrillary acidic protein in mice. J Neurosci 2002; 22: 6972–6979.
Ferrarese R, GRt Harsh, Yadav AK, Bug E, Maticzka D, Reichardt W et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest 2014; 124: 2861–2876.
Olshen AB, Venkatraman ES, Lucito R, Wigler M . Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004; 5: 557–572.
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G . GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12: R41.
Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B . Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 2001; 15: 2069–2082.
Dionyssiou MG, Salma J, Bevzyuk M, Wales S, Zakharyan L, McDermott JC . Kruppel-like factor 6 (KLF6) promotes cell proliferation in skeletal myoblasts in response to TGFbeta/Smad3 signaling. Skelet Muscle 2013; 3: 7.
Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010; 463: 318–325.
Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J 2015; 34: 1612–1629.
Varghese F, Bukhari AB, Malhotra R, De A . IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One 2014; 9: e96801.
Acknowledgements
This study was supported by National Cancer Institute grant P20 CA151129-01A1/UAB Specialized Program of Research Excellence (SPORE) in Brain Cancer (M Bredel), German Cancer Aid Grant Award (107714, M Bredel), State of Alabama Investment Pool for Action (IMPACT) funds (M Bredel). We thank Jürgen Beck (University of Freiburg) for helping in preparing reagents, Maria Oberle (University of Freiburg) for help with mice brain histology, Dr Veerle Baekelandt (University of Leuven) for providing the PCHMWS-eGFP-IRES lentiviral vector, Dr Thomas Brabletz (University of Erlangen) for providing NF-κB luciferase vectors and Darren Ó hAilín (University of Freiburg) and Susanne Nozell (UAB) for editing the manuscript.
Author contributions
APM, RF, EK, FD, MH, TU and LP performed the experiments; NKT, HK and DMS performed statistical analysis, AW provided with surgical samples, MP offered support for histology analysis, GYG provided with GBM xenolines, GRH was involved in generating data that led to this project; MSC and MB designed the research study and supervised the project, APM, MSC and MB wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Masilamani, A., Ferrarese, R., Kling, E. et al. KLF6 depletion promotes NF-κB signaling in glioblastoma. Oncogene 36, 3562–3575 (2017). https://doi.org/10.1038/onc.2016.507
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.507
This article is cited by
-
KLF6 alleviates hepatic ischemia-reperfusion injury by inhibiting autophagy
Cell Death & Disease (2023)
-
Identification and characterization of a BRAF fusion oncoprotein with retained autoinhibitory domains
Oncogene (2020)
-
Oligonucleotide Therapeutics as a New Class of Drugs for Malignant Brain Tumors: Targeting mRNAs, Regulatory RNAs, Mutations, Combinations, and Beyond
Neurotherapeutics (2019)
-
Genetic heterogeneity of primary lesion and metastasis in small intestine neuroendocrine tumors
Scientific Reports (2018)
-
Genome-wide expression profiling of glioblastoma using a large combined cohort
Scientific Reports (2018)