Abstract
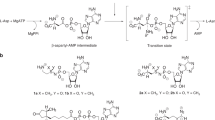
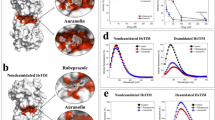
Type 2 transglutaminase (TG2) is an important cancer stem cell survival protein that exists in open and closed conformations. The major intracellular form is the closed conformation that functions as a GTP-binding GTPase and is required for cancer stem cell survival. However, at a finite rate, TG2 transitions to an open conformation that exposes the transamidase catalytic site involved in protein–protein crosslinking. The activities are mutually exclusive, as the closed conformation has GTP binding/GTPase activity, and the open conformation transamidase activity. We recently showed that GTP binding, but not transamidase activity, is required for TG2-dependent cancer stem cell invasion, migration and tumour formation. However, we were surprised that transamidase site-specific inhibitors reduce cancer stem cell survival. We now show that compounds NC9, VA4 and VA5, which react exclusively at the TG2 transamidase site, inhibit both transamidase and GTP-binding activities. Transamidase activity is inhibited by direct inhibitor binding at the transamidase site, and GTP binding is blocked because inhibitor interaction at the transamidase site locks the protein in the extended/open conformation to disorganize/inactivate the GTP binding/GTPase site. These findings suggest that transamidase site-specific inhibitors can inhibit GTP binding/signalling by driving a conformation change that disorganizes the TG2 GTP binding to reduce TG2-dependent signalling, and that drugs designed to target this site may be potent anti-cancer agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
03 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-01709-5
References
Griffin M, Casadio R, Bergamini CM . Transglutaminases: nature’s biological glues. Biochem J 2002; 368: 377–396.
Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GV et al. Transglutaminase regulation of cell function. Physiol Rev 2014; 94: 383–417.
Nakaoka H, Perez DM, Baek KJ, Das T, Husain A, Misono K et al. Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science 1994; 264: 1593–1596.
Begg GE, Carrington L, Stokes PH, Matthews JM, Wouters MA, Husain A et al. Mechanism of allosteric regulation of transglutaminase 2 by GTP. Proc Natl Acad Sci USA 2006; 103: 19683–19688.
Begg GE, Holman SR, Stokes PH, Matthews JM, Graham RM, Iismaa SE . Mutation of a critical arginine in the GTP-binding site of transglutaminase 2 disinhibits intracellular cross-linking activity. J Biol Chem 2006; 281: 12603–12609.
Gundemir S, Johnson GV . Intracellular localization and conformational state of transglutaminase 2: implications for cell death. PLoS One 2009; 4: e6123.
Pinkas DM, Strop P, Brunger AT, Khosla C . Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol 2007; 5: e327.
Gundemir S, Colak G, Tucholski J, Johnson GV . Transglutaminase 2: a molecular Swiss army knife. Biochim Biophys Acta 2012; 1823: 406–419.
Jang TH, Lee DS, Choi K, Jeong EM, Kim IG, Kim YW et al. Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site. PLoS One 2014; 9: e107005.
Stamnaes J, Pinkas DM, Fleckenstein B, Khosla C, Sollid LM . Redox regulation of transglutaminase 2 activity. J Biol Chem 2010; 285: 25402–25409.
Kiraly R, Csosz E, Kurtan T, Antus S, Szigeti K, Simon-Vecsei Z et al. Functional significance of five noncanonical Ca2+-binding sites of human transglutaminase 2 characterized by site-directed mutagenesis. FEBS J 2009; 276: 7083–7096.
Bergamini CM . GTP modulates calcium binding and cation-induced conformational changes in erythrocyte transglutaminase. FEBS Lett 1988; 239: 255–258.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Cariati M, Purushotham AD . Stem cells and breast cancer. Histopathology 2008; 52: 99–107.
Clevers H . The cancer stem cell: premises, promises and challenges. Nat Med 2011; 17: 313–319.
Eckert RL, Fisher ML, Grun D, Adhikary G, Xu W, Kerr C . Transglutaminase is a tumor cell and cancer stem cell survival factor. Mol Carcinog 2015; 54: 947–958.
Adhikary G, Grun D, Kerr C, Balasubramanian S, Rorke EA, Vemuri M et al. Identification of a population of epidermal squamous cell carcinoma cells with enhanced potential for tumor formation. PLoS One 2013; 8: e84324.
Fisher ML, Adhikary G, Xu W, Kerr C, Keillor JW, Eckert RL . Type II transglutaminase stimulates epidermal cancer stem cell epithelial-mesenchymal transition. Oncotarget 2015; 6: 20525–20539.
Fisher ML, Keillor JW, Xu W, Eckert RL, Kerr C . Transglutaminase is required for epidermal squamous cell carcinoma stem cell survival. Mol Cancer Res 2015; 13: 1083–1094.
Herman JF, Mangala LS, Mehta K . Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene 2006; 25: 3049–3058.
Kumar A, Xu J, Brady S, Gao H, Yu D, Reuben J et al. Tissue transglutaminase promotes drug resistance and invasion by inducing mesenchymal transition in mammary epithelial cells. PLoS One 2010; 5: e13390.
Kumar A, Xu J, Sung B, Kumar S, Yu D, Aggarwal BB et al. Evidence that GTP-binding domain but not catalytic domain of transglutaminase 2 is essential for epithelial-to-mesenchymal transition in mammary epithelial cells. Breast Cancer Res 2012; 14: R4.
Griffin M, Mongeot A, Collighan R, Saint RE, Jones RA, Coutts IG et al. Synthesis of potent water-soluble tissue transglutaminase inhibitors. Bioorg Med Chem Lett 2008; 18: 5559–5562.
Keillor JW, Apperley KY, Akbar A . Inhibitors of tissue transglutaminase. Trends Pharmacol Sci 2015; 36: 32–40.
Keillor JW, Clouthier CM, Apperley KY, Akbar A, Mulani A . Acyl transfer mechanisms of tissue transglutaminase. Bioorg Chem 2014; 57: 186–197.
Prime ME, Andersen OA, Barker JJ, Brooks MA, Cheng RK, Toogood-Johnson I et al. Discovery and structure-activity relationship of potent and selective covalent inhibitors of transglutaminase 2 for Huntington’s disease. J Med Chem 2012; 55: 1021–1046.
Siegel M, Khosla C . Transglutaminase 2 inhibitors and their therapeutic role in disease states. Pharmacol Ther 2007; 115: 232–245.
Yuan L, Siegel M, Choi K, Khosla C, Miller CR, Jackson EN et al. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene 2007; 26: 2563–2573.
Keillor JW, Chica RA, Chabot N, Vinci V, Pardin C, Fortin E et al. The bioorganic chemistry of transglutaminase — from mechanism to inhibition and engineering. Can J Chem 2008; 86: 271–276.
Caron NS, Munsie LN, Keillor JW, Truant R . Using FLIM-FRET to measure conformational changes of transglutaminase type 2 in live cells. PLoS One 2012; 7: e44159.
Mehta K . Biological and therapeutic significance of tissue transglutaminase in pancreatic cancer. Amino Acids 2009; 36: 709–716.
Chen Y, Mills JD, Periasamy A . Protein localization in living cells and tissues using FRET and FLIM. Differentiation 2003; 71: 528–541.
Lakowicz JR, Szmacinski H, Nowaczyk K, Berndt KW, Johnson M . Fluorescence lifetime imaging. Anal Biochem 1992; 202: 316–330.
Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML . Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci USA 1992; 89: 1271–1275.
Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW . Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal 2006; 12: 238–254.
Wallrabe H, Periasamy A . Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol 2005; 16: 19–27.
Rizzo MA, Springer GH . Granada B and Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 2004; 22: 445–449.
Keillor JW, Chabot N, Roy I, Mulani A, Leogane O, Pardin C . Irreversible inhibitors of tissue transglutaminase. Adv Enzymol Relat Areas Mol Biol 2011; 78: 415–447.
Pardin C, Roy I, Lubell WD, Keillor JW . Reversible and competitive cinnamoyl triazole inhibitors of tissue transglutaminase. Chem Biol Drug Des 2008; 72: 189–196.
Jans R, Sturniolo MT, Eckert RL . Localization of the TIG3 transglutaminase interaction domain and demonstration that the amino-terminal region is required for TIG3 function as a keratinocyte differentiation regulator. J Invest Dermatol 2008; 128: 517–529.
Oliverio S, Amendola A, Di Sano F, Farrace MG, Fesus L, Nemes Z et al. Tissue transglutaminase-dependent posttranslational modification of the retinoblastoma gene product in promonocytic cells undergoing apoptosis. Mol Cell Biol 1997; 17: 6040–6048.
Sturniolo MT, Chandraratna RA, Eckert RL . A novel transglutaminase activator forms a complex with type 1 transglutaminase. Oncogene 2005; 24: 2963–2972.
Sturniolo MT, Dashti SR, Deucher A, Rorke EA, Broome AM, Chandraratna RA et al. A novel tumor suppressor protein promotes keratinocyte terminal differentiation via activation of type I transglutaminase. J Biol Chem 2003; 278: 48066–48073.
Clouthier CM, Mironov GG, Okhonin V, Berezovski MV, Keillor JW . Real-time monitoring of protein conformational dynamics in solution using kinetic capillary electrophoresis. Angew Chem Int Ed Engl 2012; 51: 12464–12468.
Pardin C, Roy I, Chica RA, Bonneil E, Thibault P, Lubell WD et al. Photolabeling of tissue transglutaminase reveals the binding mode of potent cinnamoyl inhibitors. Biochemistry 2009; 48: 3346–3353.
Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE . Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 1988; 106: 761–771.
Liu S, Cerione RA, Clardy J . Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc Natl Acad Sci USA 2002; 99: 2743–2747.
Agnihotri N, Kumar S, Mehta K . Tissue transglutaminase as a central mediator in inflammation-induced progression of breast cancer. Breast Cancer Res 2013; 15: 202.
Kumar A, Gao H, Xu J, Reuben J, Yu D, Mehta K . Evidence that aberrant expression of tissue transglutaminase promotes stem cell characteristics in mammary epithelial cells. PLoS One 2011; 6: e20701.
Fu J, Yang QY, Sai K, Chen FR, Pang JC, Ng HK et al. TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro Oncol 2013; 15: 1353–1365.
Cao L, Shao M, Schilder J, Guise T, Mohammad KS, Matei D . Tissue transglutaminase links TGF-beta, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene 2012; 31: 2521–2534.
Badarau E, Collighan RJ, Griffin M . Recent advances in the development of tissue transglutaminase (TG2) inhibitors. Amino Acids 2013; 44: 119–127.
Pavlyukov MS, Antipova NV, Balashova MV, Shakhparonov MI . Detection of transglutaminase 2 conformational changes in living cell. Biochem Biophys Res Commun 2012; 421: 773–779.
Eckert RL, Sturniolo MT, Jans R, Kraft CA, Jiang H, Rorke EA . TIG3: a regulator of type I transglutaminase activity in epidermis. Amino Acids 2009; 36: 739–746.
Adhikary G, Grun D, Balasubramanian S, Kerr C, Huang J, Eckert R . Survival of skin cancer stem cells requires the Ezh2 polycomb group protein. Carcinogenesis 2015; 36: 800–810.
Acknowledgements
This work was supported by National Institutes of Health grants R01-CA184027 and R01-CA131074 to Richard Eckert.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Kerr, C., Szmacinski, H., Fisher, M. et al. Transamidase site-targeted agents alter the conformation of the transglutaminase cancer stem cell survival protein to reduce GTP binding activity and cancer stem cell survival. Oncogene 36, 2981–2990 (2017). https://doi.org/10.1038/onc.2016.452
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.452
This article is cited by
-
Transglutaminase 2 associated with PI3K and PTEN in a membrane-bound signalosome platform blunts cell death
Cell Death & Disease (2023)
-
A Controlled Transcription-Driven Light-Up Aptamer Amplification for Nucleoside Triphosphate Detection
BioChip Journal (2023)
-
Probing tissue transglutaminase mediated vascular smooth muscle cell aging using a novel transamidation-deficient Tgm2-C277S mouse model
Cell Death Discovery (2021)
-
Proteomic sift through serum and endometrium profiles unraveled signature proteins associated with subdued fertility and dampened endometrial receptivity in women with polycystic ovary syndrome
Cell and Tissue Research (2020)
-
Involvement of non-coding RNAs and transcription factors in the induction of Transglutaminase isoforms by ATRA
Amino Acids (2019)