Abstract
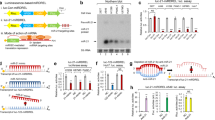
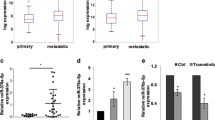
KRas is mutated in a significant number of human cancers and so there is an urgent therapeutic need to target KRas signalling. To target KRas in lung cancers we used a systems approach of integrating a genome-wide miRNA screen with patient-derived phospho-proteomic signatures of the KRas downstream pathway, and identified miR-193a-3p, which directly targets KRas. Unique aspects of miR-193a-3p biology include two functionally independent target sites in the KRas 3′UTR and clinically significant correlation between miR-193a-3p and KRas expression in patients. Rescue experiments with mutated KRas 3’UTR showed very significantly that the anti-tumour effect of miR-193a-3p is via specific direct targeting of KRas and not due to other targets. Ex vivo and in vivo studies utilizing nanoliposome packaged miR-193a-3p demonstrated significant inhibition of tumour growth, circulating tumour cell viability and decreased metastasis. These studies show the broader applicability of using miR-193a-3p as a therapeutic agent to target KRas-mutant cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Jancik S, Drabek J, Radzioch D, Hajduch M . Clinical relevance of KRAS in human cancers. J Biomed Biotechnol 2010; 2010: 150960.
Der CJ, Cooper GM . Altered gene products are associated with activation of cellular rasK genes in human lung and colon carcinomas. Cell 1983; 32: 201–208.
Bos JL . ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689.
Uberall I, Kolar Z, Trojanec R, Berkovcova J, Hajduch M . The status and role of ErbB receptors in human cancer. Exp Mol Pathol 2008; 84: 79–89.
Katzel JA, Fanucchi MP, Li Z . Recent advances of novel targeted therapy in non-small cell lung cancer. J Hematol Oncol 2009; 2: 2.
Agarwal A, Eide CA, Harlow A, Corbin AS, Mauro MJ, Druker BJ et al. An activating KRAS mutation in imatinib-resistant chronic myeloid leukemia. Leukemia 2008; 22: 2269–2272.
Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008; 26: 374–379.
Rosell R, Gonzalez-Larriba JL, Alberola V, Molina F, Monzo M, Benito D et al. Single-agent paclitaxel by 3-hour infusion in the treatment of non-small cell lung cancer: links between p53 and K-ras gene status and chemosensitivity. Semin Oncol 1995; 22 (6 Suppl 14): 12–18.
Grana TM, Rusyn EV, Zhou H, Sartor CI, Cox AD . Ras mediates radioresistance through both phosphatidylinositol 3-kinase-dependent and Raf-dependent but mitogen-activated protein kinase/extracellular signal-regulated kinase kinase-independent signaling pathways. Cancer Res 2002; 62: 4142–4150.
Wu SY, Lopez-Berestein G, Calin GA, Sood AK . RNAi therapies: drugging the undruggable. Sci Transl Med 2014; 6: 240ps7.
Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PloS One 2012; 7: e32068.
Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med 2012; 4: 206–217.
Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun 2014; 5: 3887.
Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal 2014; 7: ra42.
Kalinichenko VV, Kalin TV . Is there potential to target FOXM1 for ‘undruggable’ lung cancers? Expert Opin Ther Targets 2015; 19: 865–867.
Hong X, Nguyen HT, Chen Q, Zhang R, Hagman Z, Voorhoeve PM et al. Opposing activities of the Ras and Hippo pathways converge on regulation of YAP protein turnover. EMBO J 2014; 33: 2447–2457.
Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–550.
Moss TJ, Luo Z, Seviour EG, Sehgal V, Lu Y, Hill SM et al. Genome-wide perturbations by miRNAs map onto functional cellular pathways, identifying regulators of chromatin modifiers. Npj Syst Biol Appl 2015; 1: 15001.
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F et al. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene 2015; 35: 801.
Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH . starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 2011; 39(Database issue): D202–D209.
Lv L, Deng H, Li Y, Zhang C, Liu X, Liu Q et al. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis 2014; 5: e1402.
Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res 2012; 18: 1619–1629.
Iliopoulos D, Rotem A, Struhl K . Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res 2011; 71: 5144–5153.
Komurov K, Dursun S, Erdin S, Ram PT . NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics 2012; 13: 282.
Komurov K, White M, Ram PT . Use of data-biased random walks on graphs for the retrieval of context-specific networks from genomic data. PLoS Comput Biol 2010; 6: pii: e1000889.
Mishra DK, Thrall MJ, Baird BN, Ott HC, Blackmon SH, Kurie JM et al. Human lung cancer cells grown on acellular rat lung matrix create perfusable tumor nodules. Ann Thorac Surg 2012; 93: 1075–1081.
Mishra DK, Creighton CJ, Zhang Y, Chen F, Thrall MJ, Kim MP . Ex vivo four-dimensional lung cancer model mimics metastasis. Ann Thorac Surg 2015; 99: 1149–1156.
Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH . Delivery of RNA interference. Cell Cycle 2006; 5: 2103–2109.
Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013; 10: 1046–1047.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Cancer Genome Atlas N.. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337.
Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615.
Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–525.
Supek F, Bosnjak M, Skunca N, Smuc T . REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One 2011; 6: e21800.
Mishra DK, Sakamoto JH, Thrall MJ, Baird BN, Blackmon SH, Ferrari M et al. Human lung cancer cells grown in an ex vivo 3D lung model produce matrix metalloproteinases not produced in 2D culture. PloS One 2012; 7: e45308.
Acknowledgements
All the data reported in the paper are presented. The microarray data are available from GEO under the accession number GSE73194. The work presented here was funded by NIH/NCI ICBP grant U54-CA112970 (Project 2 PI-JWG, Project 4 Co-PI’s PTR and GBM), VS was funded by the CCBTP training grant from the CPRIT, and grants from the NIH (UH2 TR000943), the RGK Foundation, and CPRIT RP110595 (AKS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Seviour, E., Sehgal, V., Mishra, D. et al. Targeting KRas-dependent tumour growth, circulating tumour cells and metastasis in vivo by clinically significant miR-193a-3p. Oncogene 36, 1339–1350 (2017). https://doi.org/10.1038/onc.2016.308
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.308
This article is cited by
-
Emerging role of non-coding RNAs in the regulation of KRAS
Cancer Cell International (2022)
-
Whole exome and transcriptome sequencing reveal clonal evolution and exhibit immune-related features in metastatic colorectal tumors
Cell Death Discovery (2021)
-
RETRACTED ARTICLE: Downregulation of miR-196-5p Induced by Hypoxia Drives Tumorigenesis and Metastasis in Hepatocellular Carcinoma
Hormones and Cancer (2019)
-
A model for RAS mutation patterns in cancers: finding the sweet spot
Nature Reviews Cancer (2018)
-
Engineering Bioartificial Lungs for Transplantation
Current Stem Cell Reports (2017)