Abstract
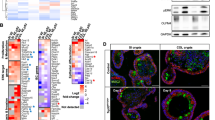
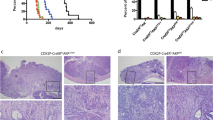
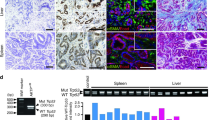
Inactivation of the adenomatous polyposis coli (APC) tumor suppressor is frequently found in colorectal cancer. Loss of APC function results in deregulation of the Wnt/β-catenin signaling pathway causing overexpression of the c-MYC oncogene. In lymphoma, both p19ARF and ribosomal proteins RPL11 and RPL5 respond to c-MYC activation to induce p53. Their role in c-MYC-driven colorectal carcinogenesis is unclear, as p19ARF deletion does not accelerate APC loss-triggered intestinal tumorigenesis. To determine the contribution of the ribosomal protein (RP)-murine double minute 2 (MDM2)-p53 pathway to APC loss-induced tumorigenesis, we crossed mice bearing MDM2C305F mutation, which disrupts RPL11- and RPL5-MDM2 binding, with Apcmin/+ mice, which are prone to intestinal tumor formation. Interestingly, loss of RP-MDM2 binding significantly accelerated colorectal tumor formation while having no discernable effect on small intestinal tumor formation. Mechanistically, APC loss leads to overexpression of c-MYC, RPL11 and RPL5 in mouse colonic tumor cells irrespective of MDM2C305F mutation. However, notable p53 stabilization and activation were observed only in Apcmin/+;Mdm2+/+ but not Apcmin/+;Mdm2C305F/C305F colon tumors. These data establish that the RP-MDM2-p53 pathway, in contrast to the p19ARF-MDM2-p53 pathway, is a critical mediator of colorectal tumorigenesis following APC loss.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ . Cancer statistics, 2009. CA 2009; 59: 225–249.
Kamangar F, Dores GM, Anderson WF . Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006; 24: 2137–2150.
Cancer Genome Atlas N.. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337.
Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991; 66: 589–600.
Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB et al. Identification of FAP locus genes from chromosome 5q21. Science 1991; 253: 661–665.
Kinzler KW, Vogelstein B . Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170.
Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 1991; 253: 665–669.
Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997; 275: 1787–1790.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT et al. Identification of c-MYC as a target of the APC pathway. Science 1998; 281: 1509–1512.
Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 2004; 18: 1385–1390.
van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002; 111: 241–250.
Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR et al. Myc deletion rescues Apc deficiency in the small intestine. Nature 2007; 446: 676–679.
Dang CV . MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 2013; 3: pii: a014217.
Eilers M, Eisenman RN . Myc's broad reach. Genes Dev 2008; 22: 2755–2766.
Sabo A, Amati B . Genome recognition by MYC. Cold Spring Harb Perspect Med 2014; 4: pii: a014191.
Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F . The c-Myc target gene network. Semin Cancer Biol 2006; 16: 253–264.
Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PloS One 2011; 6: e26057.
Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 2006; 25: 130–138.
Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene 2005; 24: 520–524.
Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 2001; 29: 397–406.
Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV . An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 2003; 4: R69.
Hoffman B, Liebermann DA . Apoptotic signaling by c-MYC. Oncogene 2008; 27: 6462–6472.
Hill RM, Kuijper S, Lindsey JC, Petrie K, Schwalbe EC, Barker K et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015; 27: 72–84.
Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985; 318: 533–538.
Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW . INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999; 13: 2670–2677.
Halberg RB, Katzung DS, Hoff PD, Moser AR, Cole CE, Lubet RA et al. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc Natl Acad Sci USA 2000; 97: 3461–3466.
Honda R, Tanaka H, Yasuda H . Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420: 25–27.
Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53 1993.
Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W . Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Mol Cell 2013; 51: 46–56.
Zhang Y, Xiong Y, Yarbrough WG . ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998; 92: 725–734.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998; 12: 2424–2433.
Kim TH, Leslie P, Zhang Y . Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 2014; 5: 860–871.
Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH . Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003; 3: 577–587.
Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 2010; 18: 231–243.
Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL . Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 1999; 13: 2658–2669.
Meng X, Carlson NR, Dong J, Zhang Y . Oncogenic c-Myc-induced lymphomagenesis is inhibited non-redundantly by the p19Arf-Mdm2-p53 and RP-Mdm2-p53 pathways. Oncogene 2015; 34: 5709–5717.
Gibson SL, Boquoi A, Chen T, Sharpless NE, Brensinger C, Enders GH . p16(Ink4a) inhibits histologic progression and angiogenic signaling in min colon tumors. Cancer Biol Ther 2005; 4: 1389–1394.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–265.
Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y . Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol 2007; 27: 1056–1068.
Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 2000; 97: 3260–3265.
Menssen A, Hermeking H . Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA 2002; 99: 6274–6279.
Toshiyuki M, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature 2011; 470: 409–413.
Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 2013; 23: 93–106.
Moser AR, Pitot HC, Dove WF . A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990; 247: 322–324.
Moser AR, Dove WF, Roth KA, Gordon JI . The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol 1992; 116: 1517–1526.
Feng Y, Sentani K, Wiese A, Sands E, Green M, Bommer GT et al. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic APC inactivation. Am J Pathol 2013; 183: 493–503.
Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J et al. Absence of nucleolar disruption after impairment of 40 S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol 2009; 11: 501–508.
Fumagalli S, Ivanenkov VV, Teng T, Thomas G . Suprainduction of p53 by disruption of 40 S and 60 S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 2012; 26: 1028–1040.
Littlewood TD, Kreuzaler P, Evan GI . All things to all people. Cell 2012; 151: 11–13.
Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 2008; 14: 447–457.
van Riggelen J, Yetil A, Felsher DW . MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10: 301–309.
Acknowledgements
This research was supported by grants from the National Institutes of Health (CA127770, CA 100302 and CA167637) and Natural Science Foundation of China (NSFC) to YZ, and the National Institute of General Medical Sciences (5T32 GM007092) to NRT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Liu, S., Tackmann, N., Yang, J. et al. Disruption of the RP-MDM2-p53 pathway accelerates APC loss-induced colorectal tumorigenesis. Oncogene 36, 1374–1383 (2017). https://doi.org/10.1038/onc.2016.301
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.301
This article is cited by
-
U2AF2-SNORA68 promotes triple-negative breast cancer stemness through the translocation of RPL23 from nucleoplasm to nucleolus and c-Myc expression
Breast Cancer Research (2024)
-
Ribosome biogenesis in disease: new players and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Loss of RPS27a expression regulates the cell cycle, apoptosis, and proliferation via the RPL11-MDM2-p53 pathway in lung adenocarcinoma cells
Journal of Experimental & Clinical Cancer Research (2022)
-
Ubiquitin ligases: guardians of mammalian development
Nature Reviews Molecular Cell Biology (2022)
-
Targeting AKT with costunolide suppresses the growth of colorectal cancer cells and induces apoptosis in vitro and in vivo
Journal of Experimental & Clinical Cancer Research (2021)