Abstract
The transcription factor nuclear factor-κB (NF-κB) has important roles for tumorigenesis, but how it regulates cancer stem cells (CSCs) remains largely unclear. We identified insulin-like growth factor 2 (IGF2) is a key target of NF-κB activated by HER2/HER3 signaling to form tumor spheres in breast cancer cells. The IGF2 receptor, IGF1 R, was expressed at high levels in CSC-enriched populations in primary breast cancer cells. Moreover, IGF2-PI3K (IGF2-phosphatidyl inositol 3 kinase) signaling induced expression of a stemness transcription factor, inhibitor of DNA-binding 1 (ID1), and IGF2 itself. ID1 knockdown greatly reduced IGF2 expression, and tumor sphere formation. Finally, treatment with anti-IGF1/2 antibodies blocked tumorigenesis derived from the IGF1Rhigh CSC-enriched population in a patient-derived xenograft model. Thus, NF-κB may trigger IGF2-ID1-IGF2-positive feedback circuits that allow cancer stem-like cells to appear. Then, they may become addicted to the circuits. As the circuits are the Achilles’ heels of CSCs, it will be critical to break them for eradication of CSCs.
Similar content being viewed by others
Introduction
Cancer stem cells (CSCs) are thought be responsible for tumor, recurrence and drug resistance. Target therapies against CSCs are still unmet medial needs.1 Tumor tissues are comprised of a wide variety of heterogeneous cell types and are thought to be maintained in a hierarchical organization involving a relatively small number of CSCs and higher numbers of dividing progenitor cells and differentiated tumor cells, similar to how normal tissues are derived from tissue-specific stem cells.1, 2, 3, 4, 5 CSCs represent a distinct cell population with the capacity for self-renewal that can prospectively be isolated. Several properties of CSCs have been described, and cancer cells that exhibit some CSC properties have been detected in many solid tumors, including breast cancer.3, 6
CSCs are maintained by their surrounding tumor microenvironment, known as the CSC niche.7 These CSC niche cells are composed of various types, including tumor cells, which are the progeny of the CSCs. CSCs may survive after systemic treatment owing to protection by the niche cells, causing recurrence or drug resistance. Mathematical models also support the concept that a small number of CSCs are maintained in the tumor tissues, although the molecular mechanisms remain largely unclear.8 Thus, there is an urgent need for identification of key mechanisms that have important roles for maintenance of the stemness; these mechanisms could prove to be the Achilles’ heel of CSCs, and provide a rationale for development of novel molecular targeted therapies to eradicate tumors.
Emerging evidence suggests that there is a chronic inflammatory microenvironment in the CSC niche.7, 9 It appears that the activity of nuclear factor-κB (NF-κB), a key transcription factor for inflammation, is increased in the tumor microenvironment.10 The increased activity of NF-κB appears to have important roles for endowing cancer cells with the stem-like properties.10, 11, 12, 13, 14 NF-κB is a heterodimer complex that binds to IκB in an inactive state in the cytoplasm.15 It appears that HER2/HER3, a heterodimer of members of the epidermal growth factor (EGF) receptor family, activates the phosphatidyl inositol 3 kinase (PI3K)/Akt pathway, leading to phosphorylation of IκB in breast cancer cells.16 Then, phosphorylated IκB undergoes ubiquitylation/degradation and the released NF-κB heterodimer is transported to the nucleus for transcriptional activation to increase the stemness of breast cancer cells. The key transcriptional targets of NF-κB to increase the stemness of breast cancer cells remain largely unclear.
The ability for in vitro tumor sphere formation has been established as a property of CSCs.17, 18 Tumor spheres are floating cell aggregates that are produced when cancer cells are cultured in defined sphere culture medium (SCM) containing a cocktail of growth factors and hormones. Epithelial cells do not survive in suspension; however, cells with stem-like properties are thought to survive and be able to divide in suspension.19 As it appears that cancer cell lines may survive in suspension because of immortalization, cancer cell lines may have limited usefulness for analyzing tumor sphere-forming ability. It is thus important to use early-passage patient-derived primary cancer cells. We previously reported that heregulin (HRG), a ligand for HER3, can strongly stimulate tumor sphere formation as the sole factor in patient-derived breast cancer cells through HER2/HER3-PI3K/Akt-NF-κB pathway.16
Insulin-like growth factor 2 (IGF2) is a member of the insulin family. IGF2 binds to IGF1 receptor (IGF-1Rs) homodimers and to IGF1 R and insulin receptor (IR) heterodimers, resulting in PI3K activation, whereas insulin binds to IR homodimers.20 Although insulin expression is confined to pancreatic β-cells, overexpression of IGF2 has been reported in many types of malignancies. IGF1 R signaling appears to confer resistance to radiation to glioma stem cells.21
Inhibitor of DNA-binding 1 (ID1) is a member of the ID family of proteins, which are known to control transcription.22, 23 ID proteins bind to basic helix–loop–helix transcription factors that have roles in the negative regulation of cell differentiation, leading to maintenance of stemness.24, 25, 26 ID proteins have been reported to be aberrantly expressed in many types of malignancies.27
To identify key downstream targets of NF-κB that are activated via the HER2/HER3-PI3K-Akt pathway and that are important to increase the stemness of breast cancer cells, we systematically analyzed gene expression profiles over time in breast cancer cells stimulated with heregulin (HRG). We identified many candidate molecules, including many cytokines/chemokines, many of which may have roles in the interaction between CSCs and their niche. We identified the IGF2-ID1-IGF2-positive feedback circuit as a critical mechanism by which the stemness may be increased and maintained in cancer cells. Furthermore, we showed that anti-IGF2/1 treatment completely blocked tumorigenesis in cells derived from an IGF1Rhigh CSC-enriched population in a patient-derived xenograft model. Once the stemness circuit is established, the cancer cells with stem-like properties may become addicted to similar mechanisms for the maintenance of stemness. We thus provide a rationale for development of novel therapies that break the stemness circuit, the Achilles’ heel of cancer stem-like cells. Importantly, it appears that the cancer stem-like cells are addicted to the circuit, irrespective of the breast cancer subtype: luminal A, luminal B, HER2-positive or triple-negative subtype.
Results
IGF2 is a critical downstream target of the HER2/HER3-PI3K-NF-κB pathway for tumor sphere formation of patient-derived primary breast cancer cells
To identify genes that are potentially upregulated downstream of HER2/HER3-PI3K-NF-κB pathway, we examined the gene expression profiles in MCF7 breast cancer cells stimulated with HRG in the absence or presence of DHMEQ or LY294002, which are specific inhibitors of NF-κB and PI3K, respectively. Because changes in gene expression levels over time are highly dynamic following stimulation with growth factors,28, 29 we examined many time points, every hour for up to 16 h after stimulation. Our criteria for candidate genes were that their expression levels were increased by stimulation with HRG and inhibited by additional treatment with the above-mentioned inhibitors. Because we obtained data at many time points, we considered differences in expression levels to be significant even when <1.5-fold at each time point. Several cytokines/chemokines and their receptors were identified among the upregulated genes. When we focused on early time points (up to 6 h after stimulation), the tendency for upregulation of these genes was more obvious (Figure 1a and Supplementary Figure 1). These results suggest that cytokines/chemokines and their receptors are directly regulated by the HER2/HER3-PI3K-NF-κB pathway. In total, 100 candidate genes, including the above-mentioned genes, were obtained.
IGF2 stimulates the sphere formation of most of the patient-derived primary breast cancer cells. (a) A Venn diagram of genes encoding cytokines/chemokines downstream of HER2/HER3-PI3K-NF-κB pathway that were upregulated at early time points (up to 6 h) and/or throughout the time course (0–16 h) after HRG treatment. (b and e) Representative phase contrast images of primary culture of tumor spheres derived from breast cancer patients. The cells were incubated with or without IGF2 (200 ng/ml), AR (20 ng/ml) or SCM (EGF/bFGF/B27). Scale bar: 100 μm. (c and d) Induction of spheres by IGF2 in different samples of breast cancer patients (c) and in different cancer cell lines (d). (f and g) Induction of spheres by AR compared with EGF (20 ng/ml) (f) or by IL-20 (200 ng/ml) compared with SCM (g) in patient-derived breast cancer cells. For (c–g), the spheres were counted and the percentage of sphere-forming cells were determined for each treatment group (n=4). **P<0.01; ***P<0.001.
We focused IGF2, amphiregulin (AR) and interleukin-20 (IL-20). IGF2 was identified as being upregulated at both early time points and continuously throughout the time course (Figure 1a and Supplementary Figure 1). We validated the ability of IGF2 to induce sphere formation of patient-derived primary breast cancer cells, and then compared its sphere-inducing ability with that of SCM. When IGF2 was the sole factor present, it induced sphere formation in all patient-derived primary breast cancer cell types in which SCM was able to induce spheres, including cells from luminal subtype and triple-negative subtypes (15/15 samples) (Figures 1b and c and Supplementary Table 1). IGF2 also stimulated sphere formation in many breast cancer cell lines, including the luminal-type MCF7 and T47D, basal-type BT20 and HER2-positive-type BT474 cell lines (Figure 1d). However, IGF2 modestly inhibited cell growth activity in MCF7 cells under adherent culture conditions (Supplementary Figure 2).
AR is a ligand for the EGF receptor.30 When AR was the sole factor present, it induced sphere formation in several patient-derived primary cancer cells, including cells of the luminal and triple-negative subtypes, but could not induce sphere formation in all of the cases in which SCM was able to induce spheres (2/5 samples) (Figures 1e and f). AR was able to induce sphere formation of many breast cancer cell lines, including luminal-type MCF7, basal-type BT20, HCC1143 and HCC1954 lines (Supplementary Figures 3a and b). AR-induced sphere formation more strongly than did EGF.
We next studied the cytokine IL-20. When IL-20 was the sole factor present, it did not significantly induce sphere formation in any patient-derived primary cancer cells examined (>5 samples, including luminal and triple-negative subtypes), whereas SCM induced spheres (Figure 2g and data not shown). However, IL-20 induced sphere formation in several breast cancer cell lines, including the MCF7 and BT474 lines (Supplementary Figures 4a and b). These results indicate that IGF2 and AR, but not IL-20, promote tumor sphere formation in patient-derived primary cancer cells. The different effects of IL-20 on sphere formation in primary cancer cells and in cancer cell lines suggest that cancer cell lines are more prone to form spheres than primary cancer cells, reinforcing the limitations of cell lines.
IGF2 is a key factor for the sphere formation. (a) Reduction of HRG-induced sphere formation of MCF7 cells and of patient-derived primary breast cancer cells by treatment with an anti-IGF2-neutralizing antibody (IGF2 NAb). (b) HRG-induced sphere formation of MCF7 cells was not significantly affected by treatment with an anti-AR neutralizing antibody (AR NAb). The spheres were counted and the percentage of sphere-forming cells were determined for each treatment group (n=4). **P<0.01; ***P<0.001.
Since HRG stimulation induces production of many soluble factors, not just IGF2 or AR (Figure 1a and Supplementary Figure 1), other soluble factors may compensate during sphere formation. Another possibility is that either alone may be a critical factor for sphere formation, exceeding the effects of the others. We next analyzed sphere formation in the presence of anti-IGF2 or anti-AR antibodies. When patient-derived primary cancer cells or MCF7 cells were treated with an anti-IGF2 antibody during induction of sphere formation by stimulation with HRG, sphere formation was greatly inhibited (Figure 2a). These results indicated that HRG-induced sphere formation is dependent on IGF2 activity. In contrast, when MCF7 cells were similarly treated with an anti-AR antibody, HRG-induced sphere formation was not significantly affected (Figure 2b). As AR-induced sphere formation was markedly inhibited by treatment with the anti-AR antibody, the neutralizing activity of the anti-AR antibody remained intact, suggesting that AR induction of spheres is only a minor component of HRG-induced sphere formation. Thus, although HRG stimulation appears to induce many soluble factors, including cytokines/chemokines, IGF2 is a key downstream target of the HER2/HER3-PI3K-NF-κB pathway in HRG-induced sphere formation.
Overexpression of IGF2 increases the tumor-initiating ability of MCF7 cells
To further examine the function of IGF2 in sphere formation, we overexpressed IGF2 in MCF7 cells using a lentivirus system (Figure 3a). IGF2-overexpressing cells displayed sphere formation, even in the absence of any growth factors or hormones (Figure 3b). Sphere formation was blocked by treatment with the anti-IGF2 antibody, confirming that this activity was dependent on IGF2 (Figure 3c). Tumor-initiating ability is another criterion for CSC properties,18 and we compared the tumor-initiating ability of IGF2-expressing cells with that of vector-transduced cells. Following limiting dilution, we inoculated these cells into the mammary fat pads of immune-compromised NOD scid gamma (NSG) mice (Figure 3d). The tumor-initiating cell frequency was estimated to be much higher in IGF2-overexpressing cells compared with that in control vector-transduced cells (1122-fold; 1/309 vs 1/591 395, P=1.62e−33; Table 1). Thus, IGF2-overexpressing cells show stronger tumor-initiating activity than do vector-transduced cells.
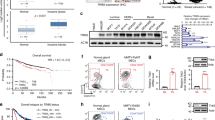
Overexpression of IGF2 increases sphere-forming activity and tumor-initiating activity and correlation of IGF2 expression with breast cancer malignancy and prognosis. (a) Immunoblotting analysis of IGF2 expression in MCF7 cells transduced with a lentiviral control (Vector) or IGF2-expressing vector (IGF2-OE). Actin was used as a loading control. (b) Representative phase contrast images of tumor spheres of the cells in (a). Scale bar: 100 μm. (c) The number of spheres of the cells in (a and b) were counted and the effect of anti-IGF2 antibody (1 mg/ml) on sphere formation of the IGF2-overexpressing cells was analyzed (n=4). (d) Representative images of tumors generated in mice injected with cells described in (a). Scale bar: 1 cm. (e) Representative images of anti-IGF2 antibody immunohistochemical staining of tissue microarray of breast cancer samples. (f) Quantification of the expression levels of IGF2 in grade I and grade II/III breast cancer tissues. (g) Kaplan–Meier plots for estimation of disease-specific survival of patients with breast cancer tissues showing low or high IGF2 gene signature. **P<0.01.
Expression of IGF2 correlates with histological malignancy, recurrence and metastasis of breast cancer
CSC activity is thought to be associated with the malignant phenotype of human breast cancer.3, 4 Histological grading of breast cancer tissues is associated with cancer malignancy, with a higher grade indicating a more aggressive and faster-growing cancer. We used tissue microarray sections of samples from primary breast cancer tumors (n=105) to examine correlations between histological grade and IGF2 immunohistochemical expression level. Among 105 cases, 90 were positive for IGF2 and 15 were negative. The IGF2-positive cases were more frequently of a high histological grade (grade II or grade III) compared with the IGF2-negative cases (P=0.0002, Fisher’s exact test) (Figures 3e and f). We compared the histological grades with major clinicopathological parameters and found that nodal involvement-positive and progesterone receptor (PR)-negative cases were significantly correlated to high histological grades (Supplementary Table 2).
We next analyzed the association between IGF2 signaling activity in cancer tissues and the prognosis of breast cancer patients. We performed DNA microarray analysis of IGF2-overexpressing MCF7 cells and control cells to identify genes in which the expression levels were positively or negatively regulated by overexpression of IGF2. Then, combined analysis of expression profiles of the MCF7 cells and publically available gene expression profiles of breast cancer tissues from the patients31 allowed us to select 545 genes in which the expression levels showed a significant correlation with IGF2 expression both in MCF cells and in patient-derived cancer tissues. We designated the 545 genes (172 positively correlated genes and 373 negatively correlated genes) as ‘IGF2 gene signature’ (Supplementary Table 3). We then measured the enrichment score of the IGF2 gene signature for each tumor from a breast cancer patient using independent gene expression profiles, and assigned the tumors to high or low IGF2 gene signature groups.32, 33, 34 When comparing the high and low IGF2 gene signature groups, the high IGF2 gene signature group displayed a significantly poorer prognosis (Figure 3g)
IGF2 maintains stemness through its receptor, the IGF1 R, which is specifically expressed at high levels in a breast CSC-enriched population
To examine the mechanisms by which IGF2 has a role in breast CSCs, we analyzed IGF1 R expression in a CD44highCD24low cell population enriched for breast CSCs.6 Fluorescence-activated cell sorting analysis indicated that IGF1 R was more strongly expressed in this CSC-enriched population compared with that in the control cell population (Figure 4a). In contrast, IR expression levels were at low levels and did not differ significantly between the two populations. We then sorted patient-derived primary breast cancer cells using the anti-IGF1 R antibody to obtain cell populations in which IGF1 R was expressed at either high or low levels. When we stimulated these cells with IGF2 or HRG for sphere formation, the IGF-1Rhigh cell population gave rise to spheres (Figures 4b and c). However, the sphere-forming ability of the IGF-1Rlow cell population was markedly low under both of these conditions. Moreover, when we inoculated these cell populations, derived from patient-derived xenografts by limiting dilution, into the mammary fat pads of NSG mice, the tumor-initiating frequency was estimated to be much higher in the IGF-1Rhigh cell population compared with that in the IGF-1Rlow cell population (131-fold; 1/14 vs 1/1840, P=1.98e−11; Table 2). These results indicate that cancer stem-like cells are enriched for IGF-1R expression.
IGF1R is predominantly expressed in CSC-enriched population and IGF1Rhigh cell population shows strong sphere-forming activity. (a) Fluorescence-activated cell sorting (FACS) analysis of freshly obtained patient-derived breast cancer cells (sample no. 5, luminal subtype). The cells were sorted according to the expression of CD44 and CD24. The CD44highCD24low CSC-enriched population and the control population were then sorted according to the expression of IGF1R or IR. (b) Representative phase contrast images of sphere formation by patient-derived breast cancer cells with high or low IGF1R, which were stimulated with or without HRG (20 ng/ml) (sample no. 6, luminal subtype). Scale bar: 100 μm. (c) Quantification of the sphere-forming activity of the cells with high or low IGF1R, which were stimulated with or without IGF2 or HRG (n=4). *P<0.05.
The IGF2-PI3K-ID1-positive feedback circuit maintains a stem cell state
Our findings raised the possibility that IGF-1R signaling stimulates a stemness pathway, leading to maintenance of CSC properties. To analyze the underlying molecular mechanisms, we next performed gene set enrichment analysis,14, 35 by comparing the gene expression profiles of IGF2-overexpressing MCF7 cells with those of vector-transduced cells. We found that several gene sets associated with stemness were enriched in IGF2-overexpressing cells compared with vector-transduced cells (Supplementary Figure 5). Among the genes upregulated in IGF2-overexpressing cells, we focused on ID1, a core stemness regulator,23 as ID1 was the sixth highest-ranked gene in the IGF2 gene signature (Supplementary Table 3). Western blotting confirmed that expression of the ID1 protein was upregulated in lentivirus-transduced IGF2-overexpressing MCF7 cells compared with control vector-transduced cells (Figure 5a). Moreover, the expression level of the ID1 protein was markedly higher in IGF-1Rhigh cells compared with that in IGF-1Rlow cells originating from patient-derived primary breast cancer cells (Figure 5b).
Expression of ID1 is induced by PI3K activity downstream of HRG and IGF2 stimulation and has important roles in tumor sphere formation. (a and b) Immunoblotting analysis of ID1 expression in lentivirus-transduced control (Vector) and IGF2-overexpressing (IGF2-OE) MCF7 cells (a) and in IGF1Rhigh and IGF1Rlow patient-derived primary breast cancer cell populations (sample no. 7, triple-negative subtype) (b). Cell lysates were immunoblotted with anti-ID1, IGF1R or actin (loading control) antibodies. (c and d) MCF7 cells were stimulated with HRG (100 ng/ml), with or without treatment with LY294002 (LY) (1 mm) or anti-IGF2 antibody (1 mg/ml) (c) or with IGF2 (200 ng/ml) with or without treatment with LY294002 (1 mm). (d) Expression of ID1 was examined using qRT–PCR. Data are expressed as fold increase relative to no treatment and means±s.d.; n=3. (e) Immunoblotting analysis of the expression of ID1 protein in MCF7 cells following stimulation with or without IGF2 (200 ng/ml). Cell lysates were immunoblotted with anti-ID1 antibodies. p-AktS473 was assayed as a positive control. Actin was blotted as a loading control. (f–j) Effects of transfection of IGF2-overexpressing MCF7 cells with siRNA for ID1 or with control siRNA were examined on: (f) expression of ID1 protein by immunoblotting, (g and h) sphere-forming activity; (g) representative phase contrast images of formed spheres. Scale bar: 100 μm. (h) The spheres were counted and the percentage of sphere-forming cells were determined for each group (n=4). (i) Immunoblotting analysis of expression of Sox2 protein in control (Vector) and IGF2-overexpressing (IGF2-OE) MCF7 cells transfected with siRNA for ID1 or control siRNA. (j) Expression of IGF2 mRNA by qRT–PCR upon stimulation with IGF2 (200 ng/ml); n=3. **P<0.01; ***P<0.001.
Quantitative real time–PCR (qRT–PCR) analysis indicated that the expression levels of ID1 mRNA gradually increased over time after stimulation of breast cancer cells with HRG (Figure 5c). Treatment with an anti-IGF2 antibody blocked the HRG-induced expression of ID1, suggesting that ID1 expression is dependent on IGF2 production stimulated by HRG. The HRG-induced increase in expression of ID1 was also blocked by treatment with the PI3K inhibitor LY294002 (Figure 5c). This result indicates that PI3K activity is required for HRG-induced expression of ID1. It is known that not only HRG but also IGF2 stimulates PI3K activity.20 We confirmed that IGF2 stimulation induced expression of ID1 transcripts (Figure 5d). When we treated cells with LY294002, the IGF2-induced expression of ID1 was greatly decreased (Figure 5d). Thus, it appears that HRG stimulates production of IGF2, which stimulates PI3K to induce ID1.
We observed that IGF2 increased ID1 protein levels (Figure 5e). To examine the biological effects of ID1, we then knocked down ID1 in breast cancer cells by means of small interfering RNA (siRNA) (Figure 5f). IGF2-stimulated sphere formation was greatly decreased in ID1-knockdown cells, as shown in Figures 5g and h. HRG-stimulated sphere formation was also decreased in ID1-knockdown cells (data not shown). Western blotting indicated that expression of Sox2, a stemness marker,36 was higher in IGF2-overexpressing MCF7 cells compared with that in control vector-transduced cells and was reduced in ID1-knockdown cells (Figure 5i). In contrast, cell growth under adherent culture conditions was not significantly affected by ID1 knockdown (Supplementary Figure 6a). Apoptosis did not occur rapidly in ID1-knockdown cells under sphere culture conditions, as assessed by fluorescence-activated cell sorting analysis of the percentage of cells in the sub-G1 stage of the cell cycle (Supplementary Figure 6b). Thus, HRG stimulation induces IGF2, and then IGF2 stimulation induces ID1 to confer CSC properties.
Finally, we examined the downstream targets of ID1 that are induced by IGF2. Interestingly, we found that IGF2-induced expression of IGF2 mRNA itself (Figure 5j; siControl). Moreover, when ID1 was knocked down, IGF2 was not able to efficiently induce IGF2 mRNA expression (Figure 5j). These results suggest that an important downstream target of the IGF2-ID1 axis is IGF2 itself. Thus, an IGF2-PI3K-ID1-IGF2-positive feedback circuit is present in cancer stem-like cells, and it maintains the stem cell state. A model of the role of the molecular mechanisms for maintenance of stemness proposed above is shown in Figure 6d.
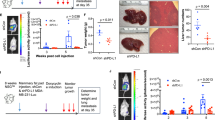
Treatment with anti-IGF1/2 antibodies inhibits tumor growth derived from IGF1Rhigh cell population. (a) IGF1Rhigh population of patient-derived breast cancer cells (sample no. 7, triple-negative subtype) were injected into the mammary fat pads of NSG mice. The mice were treated with control IgG or IGF antibodies (KM1468 and mIGF1). Tumor volume was measured every week. (b) Images of tumors generated in mice injected with patient-derived breast cancer cells. (c) Tumor weight was measured at 57 days after implantation. (d) HER2/HER3-PI3K-NF-κB pathway may trigger IGF2-PI3K-ID1-IGF2-positive feedback circuit (orange arrows) and possible PI3K-mediated feedforward circuit (blue dotted arrow) to maintain the stemness of breast cancer cells. **P<0.01.
Treatment with anti-IGF1/2 antibodies greatly inhibits initiation and growth of tumors derived from the IGF1 Rhigh cell population
To interrupt the stemness circuits in vivo, we analyzed the effects of anti-IGF1/2 antibodies on initiation and growth of tumors derived from the IGF-1Rhigh cell population. KM1468 is a neutralizing antibody against human IGF1, human IGF2 and mouse IGF2.37 We added an anti-mouse IGF1 (mIGF1) antibody to the treatment protocol to neutralize mouse IGF1, as described previously.37, 38 We inoculated 102 IGF-1Rhigh cells of patient-derived breast cancer cells (sample no. 7, triple-negative type) into the mammary fat pads of NSG mice and treated the mice with antibodies once a week (Figure 6a). Weekly monitoring showed no body weight loss in the mice treated with KM1468 (Supplementary Figure 7). Tumors of the patient-derived breast cancer cells were formed in 5/10 mice treated with control IgG after 57 days. Strikingly, no tumors were formed in mice treated with KM1468+mIGF1 (Figures 6a–c). These results indicate that anti-IGF1/2- neutralizing antibody greatly inhibits initiation and growth of tumors derived from the IGF-1Rhigh cell population of breast cancer cells in vivo.
Discussion
We have provided evidence that HER2/HER3 trigger the PI3K-NF-κB pathway, leading to production of IGF2 by breast cancer cells that serve as niche cells and by breast cancer cells with stem-like properties (Figure 6d). The resulting IGF2 binds to IGF-1R, which is specifically expressed in cancer cells with stem-like properties, and activates the PI3K pathway, leading to expression of the ID1 transcriptional regulator, which controls maintenance of stemness. In turn, ID1 leads to the production of IGF2, forming a positive feedback circuit, IGF2-IGF1R-PI3K-ID1-IGF2. Moreover, it is possible that HER2/HER3-activated PI3K not only stimulates NF-κB but also contributes to a feedforward circuit from PI3K to ID1, bypassing the NF-κB pathway, and leading to further accumulation of the ID1 protein in cancer cells with stem-like properties. These positive feedback and possible feedforward circuits may be fundamental as well as general mechanisms by which niche cancer cells and cancer cells with stem-like properties interact, and by which the stemness of breast cancer cells is maintained in cancer tissues. Once the IGF2-IGF1R-PI3K-ID1-IGF2 stemness circuit is established, CSCs may become addicted to this mechanism. Drugs targeting a few molecules in the positive feedback circuits and feedforward circuits may be effective for eradicating CSCs, and may lead to cure, by preventing recurrence.
KM1468 has been reported to significantly inhibit the growth of human prostate cancer cells in bone implanted in immunodeficient mice.37 A mixture of KM1468 and mIGF1 has also been reported to inhibit hepatic metastasis of human colon cancer cells in immunodeficient mice at low doses (0.1 μg/g mouse body weight).38 We used the same low dose of KM1468 and mIGF1 for treatment of tumors derived from an IGF1Rhigh CSC-enriched population. Treatment with a mixture of KM1468 and mIGF1 greatly inhibited tumor initiation and tumor growth. The inhibitory effects were remarkable in a patient-derived xenograft model of triple-negative breast cancer. Thus, anti-IGF1/2 therapy may considerably block recurrence and metastasis of tumors derived from breast CSCs.
Several efforts have been made to target therapeutic approaches towards IGF2 signaling, for example, with use of IGF1R-specific antibodies, anti-IGF2 antibodies and IGF1R kinase inhibitors.20, 39 Although a phase III trial of IGF1R-specific antibodies as a single agent did not produce a favorable outcome, a phase I–II trial of combination of IGF1R inhibitor and mammalian target of rapamycin inhibitor showed a favorable outcome.40 It would be important to stratify patients who are expected to respond to targeting IGF2 signaling by using predictive biomarkers, such as IGF2 expression, ideally in blood samples.
We found that the HER2/HER3-PI3K-NF-κB pathway induces many cytokines and chemokines, including AR, bone morphogenic protein 7, CXCL12, IL-20, semaphorine 3B, vascular endothelial growth factor A and platelet-derived growth factor A. These factors also appear to have roles in the interaction between cancer cells with stem-like properties and the CSC niche. As AR stimulates tumor sphere formation in some patient-derived primary breast cancer cell samples, it is possible that AR has a role in cancer stem cell function.
CSC niche cells are comprised of various cell types, viz. breast cancer cells, cancer-associated fibroblasts and immune cells, as well as endothelial cells and pericytes that form blood vessels.7 Vascular endothelial growth factor A, a typical angiogenic factor, stimulates the proliferation of endothelial cells.41 platelet-derived growth factor A stimulates the proliferation of pericytes and cancer-associated fibroblasts.42 CXCL12 is involved in many aspects of cancer biology, including cell proliferation, cell migration, CSC function and metastasis.43, 44 IL-20 may regulate immune cells.45 CSCs may manipulate all of these factors so that they can proliferate and give rise to tumors in the human body.
Proteins in the ID family, ID1–ID4, are known to be master regulators of stemness in many tissue types under normal conditions.23 A variety of molecular mechanisms strictly regulate the expression of IDs. We have provided the evidence that ID1 has important roles for the cancer stemness circuit in general.
We have very recently reported that a fusion driver gene, CD74-NRG1/HRG, found in a very minor subtype of lung adenocarcinoma confers CSC-like properties through IGF2 by in vitro study using only cell lines.46 In the present study, we have extended the analysis to find the mechanistic bases of the positive feedback IGF2 circuit by using patient-derived breast cancer cells. Moreover, we have demonstrated that the circuit is indeed the Achilles’ heel of CSCs, in general, by using patient-derived in vivo model.
Materials and methods
Primary cell culture and tumor sphere formation assay
Patient-derived tumor tissues were treated according to a previously described method.16 We previously confirmed that patient-derived tumor cells plated at 5000 cells per ml yield tumor spheres that are clonally derived from single cells.16 Sphere formation assay was performed as described previously.16 Briefly, cells were plated as single cells on ultralow attachment 24-well plates (2000–5000 cells per well). Spheres were grown in SCM containing 20 ng/ml EGF (Millipore, Billerica, MA, USA), 20 ng/ml basic fibroblast growth factor (PeproTech, Rocky Hill, NJ, USA), B27 (Gibco, Grand Island, NY, USA) and heparin (Stem Cell Technologies, Vancouver, BC, Canada) or in DMEM/F-12 medium supplemented with 20 ng/ml recombinant human HRG (R&D Systems Inc., Lille, France), 200 ng/ml recombinant human IGF2 (R&D Systems Inc.) and 20 ng/ml AR (R&D Systems Inc.) or 200 ng/ml IL-20 (R&D Systems Inc.), with or without anti-IGF2 antibody (clone S1F2; Millipore) and anti-AR antibody (R&D Systems Inc.). Spheres >75 μm in diameter were counted after 4–7 days. Sphere-forming efficiency was calculated as the ratio of the number of spheres formed to the number of cells originally plated (2000–5000 cells per well).
Identification of gene sets upregulated by the HER2/3-PI3K-NF-κB pathway at early time points up to 6 h and throughout the time course
To identify upregulated genes at early time points, we first calculated the sum of the expression values at the seven measured time points (0 and every hour up to 6 h), for each condition as below. Following this, we obtained the average gene expression value by dividing the sum by 7, for each condition. Each condition is as follows: N, no treatment; D, treatment with 1 μg/ml DHMEQ (NF-κB inhibitor); LY, treatment with 1 μm LY294002 (PI3K inhibitor) (Cell Signaling Technology, Beverly, MA, USA); H, stimulation with 100 ng/ml HRG; HD, stimulation with HRG and treatment with DHMEQ; and HLY, stimulation with HRG and treatment with LY294002. The potentially upregulated genes downtream of HER2/HER3-PI3K-NF-κB pathway were selected that satisfied the following criteria: H ≧1.25 N, H ≧1.25 D, H ≧1.25 LY, H ≧1.11 HD and H ≧1.11 HLY. We obtained data at various time points; we considered the differences in expression levels to be significant, even when they were <1.5-fold. Similarly, to identify upregulated genes throughout the time course, we calculated the average gene expression values at the 17 measured time points (0, and every hour up to 16 h), for the aforementioned conditions (GEO accession number GSE64073).
Antibodies
The following antibodies were used: anti-ID1 (C-20, 1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-IGF2 (clone S1F2, 1:5000; Millipore), anti-IGF1R (no. 9750; Cell Signaling Technology), anti-p-Akt (no. 4060; Cell Signaling Technology), anti-Sox2 (no. 3579; Cell Signaling Technology) and anti-β-actin (clone C4, 1:20 000; Millipore) antibodies.
Construction of lentiviral vectors
For construction of the lentivirus plasmid vector for expression of the IGF2 gene, the CSII-EF-MCS-IRES2-Venus vector (a kind gift from H Miyoshi, RIKEN, Tsukuba, Japan) and standard molecular biological techniques were used.47 The CSII-EF-MCS-IRES2-Venus vector is a plasmid with a gene encoding d2Venus.48 IGF2 cDNA was amplified with a primer pair encompassing the entire coding sequence of the IGF2 gene, gel-purified and was then ligated into the lentivirus plasmid vector. Lentivirus plasmid DNA (1000 ng/μl) was transfected into HEK293T cells along with packaging plasmids (pCMV-VSV-G-RSV-Rev and pCAGHIVgp) by using the Lipofectamine (Invitrogen, Karlsruhe, Germany) transfection reagent. The medium was changed after 16 h. High-titer viral stocks were prepared by ultracentrifugation.
Tissue microarray analysis
We used four different breast cancer tissue microarrays (BR951, T086b, CBA4 and CBB3) that were purchased from Biomax Inc. (http://www.biomax.us/) and Super Bio Chips Laboratories (http://www.tissue-array.com/). Tissue microarrays accommodated 105 primary breast cancer cases, totally. The demographic and clinicopathological details of patients and tumors are provided in the manufacturer's websites. Histological grading in accordance to the Nottingham grading system for breast cancer49 and immunohistochemical evaluation of IGF2 were performed by a pathologist (DM) through light microscopic observations in a blinded manner. P-values were calculated by using Fisher’s exact test.
Analysis of the prognosis of breast cancer patients of publically available gene expression profiles by using the IGF2 signature
By analysis of DNA microarray gene expression profiles of IGF2-overexpressing MCF7 cells and control cells, 670 and 422 genes were selected whose expression levels were significantly upregulated or downregulated, respectively, in IGF2-overexpressing cells compared with control cells (q-value <0.0001 by unpaired Welch's t-test and false discovery rate with the Benjamini–Hochberg procedure). Then, 545 genes were further selected and designated as ‘IGF2 gene signature’, as expression levels of each gene were significantly positively (172 genes) or negatively (373 genes) correlated with the expression levels of IGF2 in DNA microarray gene expression profiles of the breast cancer tissues, reported by Curtis et al.31 (q-value <0.01 by Pearson's product-moment correlation test and the Benjamini–Hochberg method) (Supplementary Table 3). To measure the enrichment score of the IGF2 gene signature, we performed principal component analysis of the expression profiles of these 545 genes in the expression profiles of the breast cancer patients in each data set as reported previously.50 The ‘IGF2 gene signature score’ for each patient was calculated as the first principal component. Tumors were then classified as ‘IGF2 gene signature low’ if their score was <40 percentile of the score and as ‘IGF2 gene signature high’ if their score was >60 percentile of the score. To evaluate the prognostic value of the IGF2 gene signature, we used the Kaplan–Meier method and the P-value was calculated using the log-rank test.
Anti-IGF1/2 antibody treatment in vivo
KM1468 (rat IgG2b with Fc fragment) was provided from Kyowa Hakko Kirin Co. Ltd (Tokyo, Japan). Normal goat IgG control and mouse anti-IGF1 antibody were purchased from R&D Biosystems. Patient-derived breast cancer cells (102 cells per site) were injected into the mammary fat pads of 8-week-old female immune-compromised IL2-receptor γc-dificient (NSG) mice and each antibody (0.1 μg/g mice body weight) was subsequently given intraperitoneally into mice every week. For evaluation of adverse effects due to antibody treatment, body weight of the mice was monitored every week.
Statistics
Student’s unpaired t-test was used to compare differences between two samples. For tumor sphere-forming frequency, tumor volume and tumor weight, the paired two-tailed t-test was used. Values are presented as means±s.d. Values of P<0.01–0.05 (*), P<0.001–0.01 (**) or P<0.001(***) were considered significant. Tumor-initiating frequency was calculated using the ELDA software (http://bioinf.wehi.edu.au/software/elda/index.html), provided by the Water and Eliza Hall Institute (Parkville, VIC, Australia).51
Study approval
All human breast carcinoma specimens were obtained from the University of Tokyo Hospital, Minami-Machida Hospital and Showa General Hospital. This study was approved by the institutional review boards of the Institute of Medical Science, University of Tokyo, the University of Tokyo Hospital, Minami Machida Hospital, Showa General Hospital and Kanazawa University. Written informed consent was received from all the participants before inclusion in the study.
Mice were handled according to the guidelines of the Institute of Medical Science, University of Tokyo and Kanazawa University. The experiments were approved by the committees for animal research at the Institute of Medical Science, University of Tokyo and of Kanazawa University.
Accession codes
References
Kreso A, Dick JE . Evolution of the cancer stem cell model. Cell Stem Cell 2014; 14: 275–291.
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Badve S, Nakshatri H . Breast-cancer stem cells-beyond semantics. Lancet Oncol 2012; 13: e43–e48.
Valent P, Bonnet D, De Maria R, Lapidot T, Copland M, Melo JV et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer 2012; 12: 767–775.
Gotoh N . Potential signaling pathways activated in cancer stem cells in breast cancer. In: Shostak S (eds). Cancer Stem Cells Theories and Practice InTech 2011, p 261–272.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003; 100: 3983–3988.
Plaks V, Kong N, Werb Z . The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015; 16: 225–238.
Sottoriva A, Vermeulen L, Tavare S . Modeling evolutionary dynamics of epigenetic mutations in hierarchically organized tumors. PLoS Comput Biol 2011; 7: e1001132.
Sansone P, Bromberg J . Environment, inflammation, and cancer. Curr Opin Genet Dev 2011; 21: 80–85.
Korkaya H, Liu S, Wicha MS . Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest 2011; 121: 3804–3809.
Hinohara K, Gotoh N . Inflammatory signaling pathways in self-renewing breast cancer stem cells. Curr Opin Pharmacol 2010; 10: 650–654.
Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J . NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun 2013; 4: 2299.
Wee ZN, Yatim SM, Kohlbauer VK, Feng M, Goh JY, Bao Y et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat Commun 2015; 6: 8746.
Murohashi M, Hinohara K, Kuroda M, Isagawa T, Tsuji S, Kobayashi S et al. Gene set enrichment analysis provides insight into novel signalling pathways in breast cancer stem cells. Br J Cancer 2010; 102: 206–212.
Karin M, Greten FR . NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005; 5: 749–759.
Hinohara K, Kobayashi S, Kanauchi H, Shimizu S, Nishioka K, Tsuji E et al. ErbB receptor tyrosine kinase/NF-kappaB signaling controls mammosphere formation in human breast cancer. Proc Natl Acad Sci USA 2012; 109: 6584–6589.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003; 17: 1253–1270.
Ablett MP, Singh JK, Clarke RB . Stem cells in breast tumours: are they ready for the clinic? Eur J Cancer 2012; 48: 2104–2116.
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007; 445: 111–115.
Pollak M . The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer 2012; 12: 159–169.
Osuka S, Sampetrean O, Shimizu T, Saga I, Onishi N, Sugihara E et al. IGF1 receptor signaling regulates adaptive radioprotection in glioma stem cells. Stem Cells 2013; 31: 627–640.
Sun XH, Copeland NG, Jenkins NA, Baltimore D . Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 1991; 11: 5603–5611.
Lasorella A, Benezra R, Iavarone A . The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer 2014; 14: 77–91.
Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH . Id1 but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood 2007; 110: 2351–2360.
Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA et al. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell 2010; 18: 655–668.
O'Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y et al. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell 2012; 21: 777–792.
Perk J, Gil-Bazo I, Chin Y, de Candia P, Chen JJ, Zhao Y et al. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res 2006; 66: 10870–10877.
Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One 2012; 7: e43923.
Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet 2007; 39: 503–512.
Yarden Y, Pines G . The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer 2012; 12: 553–563.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352.
Pawitan Y, Bjöhle J, Amler L, Borg AL, Egyhazi S, Hall P et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res 2005; 7: R953–R964.
Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genom 2008; 9: 239.
Schmidt M, Böhm D, von Törne C, Steiner E, Puhl A, Pilch H et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 2008; 68: 5405–5413.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Lewitzky M, Yamanaka S . Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol 2007; 18: 467–473.
Goya M, Miyamoto S, Nagai K, Ohki Y, Nakamura K, Shitara K et al. Growth inhibition of human prostate cancer cells in human adult bone implanted into nonobese diabetic/severe combined immunodeficient mice by a ligand-specific antibody to human insulin-like growth factors. Cancer Res 2004; 64: 6252–6258.
Miyamoto S, Nakamura M, Shitara K, Nakamura K, Ohki Y, Ishii G et al. Blockade of paracrine supply of insulin-like growth factors using neutralizing antibodies suppresses the liver metastasis of human colorectal cancers. Clin Cancer Res 2005; 11: 3494–3502.
Gombos A, Metzger-Filho O, Dal Lago L, Awada-Hussein A . Clinical development of insulin-like growth factor receptor—1 (IGF-1 R) inhibitors: at the crossroad? Invest New Drugs 2012; 30: 2433–2442.
Zardavas D, Baselga J, Piccart M . Emerging targeted agents in metastatic breast cancer. Nat Rev Clin Oncol 2013; 10: 191–210.
Gale NW, Yancopoulos GD . Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes Dev 1999; 13: 1055–1066.
Heldin CH . Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal 2013; 11: 97.
Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M et al. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci USA 2014; 111: E2182–E2190.
Mukherjee D, Zhao J . The Role of chemokine receptor CXCR4 in breast cancer metastasis. Am J Cancer Res 2013; 3: 46–57.
Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG . Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 2011; 29: 71–109.
Murayama T, Nakaoku T, Enari M, Nishimura T, Tominaga K, Nakata A et al. Oncogenic fusion gene CD74-NRG1 confers cancer stem cell-like properties in lung cancer through a IGF2 autocrine/paracrine circuit. Cancer Res 2016; 76: 974–983.
Miyoshi H, Smith KA, Mosier DE, Verma IM, Torbett BE . Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science 1999; 283: 682–686.
Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A . A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 2002; 20: 87–90.
AJCC Cancer Staging Manual, 7th edn. Springer: New York, NY, USA, 2010.
Bair E, Hastie T, Paul D, Tibshirani R . Prediction by surpervised principal components. J Am Stat Assoc 2006; 101: 119–137.
Hu Y, Smyth GK . ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009; 347: 70–78.
Acknowledgements
We thank H Nakauchi, Y Ishii and A Fujita for their help with flow cytometry. We are grateful to A Umezawa for his kind gift of DHMEQ. We are grateful to S Akinaga and N Shiraishi in Kyowa Hakko Kirin Co. Ltd for their kind gift of KM1468. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT (22130009), a Grant-in-Aid for Scientific Research (B) from JSPS (15H04294) and a research grant from the Ministry of Health, Labor and Welfare of Japan for the Third term Comprehensive 10-year Strategy for Cancer Control (to NG). This work was supported by Grant-in-Aid for JSPS Fellows (to KT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Tominaga, K., Shimamura, T., Kimura, N. et al. Addiction to the IGF2-ID1-IGF2 circuit for maintenance of the breast cancer stem-like cells. Oncogene 36, 1276–1286 (2017). https://doi.org/10.1038/onc.2016.293
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.293
This article is cited by
-
IGF2 is upregulated by its antisense RNA to potentiate pancreatic cancer progression
Functional & Integrative Genomics (2023)
-
Identification of a minimum number of genes to predict triple-negative breast cancer subgroups from gene expression profiles
Human Genomics (2022)
-
Insulin-like growth factor receptor signaling in tumorigenesis and drug resistance: a challenge for cancer therapy
Journal of Hematology & Oncology (2020)
-
Loss of miR-100 and miR-125b results in cancer stem cell properties through IGF2 upregulation in hepatocellular carcinoma
Scientific Reports (2020)
-
Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis
Cell Death & Differentiation (2020)