Abstract
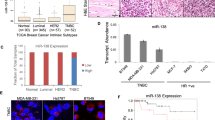
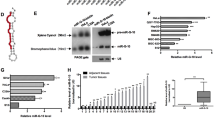
Elucidating the mechanisms involved in sensitizing radioresistant tumors to ionizing radiation (IR) treatments while minimizing injury to surrounding normal tissue is an important clinical goal. Due to their sequence-derived specificity and properties as gene regulators in IR-affected pathways, microRNAs (miRNAs) could serve as adjuvant therapeutic agents that alter cellular sensitivity to radiation treatment. To identify radiosensitizing miRNAs, we initially utilized the Caenorhabditis elegans vulval cell model, an in vivo system developed to study IR-dependent radiosensitivity as a measure of clonogenic cell death. We tested several candidate miRNA-deletion mutants post γ-irradiation and identified cel-mir-237 as a miRNA which when deleted caused animals to be more resistant to IR, whereas cel-mir-237 overexpressing strains were IR sensitive. In addition, wild-type animals downregulated cel-mir-237 levels post IR in a time-dependent manner. We identified jun-1 (JUN transcription factor homolog) as a novel target of cel-mir-237. Specifically, jun-1 transcript levels increased in wild-type animals post γ-irradiation, and loss of cel-mir-237 also resulted in higher jun-1 expression. As expected, loss of jun-1 resulted in IR sensitivity, similar to the phenotype of cel-mir-237 overexpressors. As miR-237 is the homolog of human miR-125, we validated our findings in MCF-7 and MDA-MB-231 breast cancer cell lines, which harbor lower hsa-miR-125b levels than normal human mammary epithelial cells (HMECs). Forced expression of hsa-miR-125b in these cells resulted in radiosensitivity, as seen by reduced clonogenic survival, enhanced apoptotic activity and enhanced senescence post IR. Finally, re-expression of c-JUN in MDA-MB-231 cells promoted radioresistance and abrogated miR-125-mediated radiosensitization. Our findings suggest that overexpression of cel-mir-237 and its homolog, hsa-miR-125b, functions as sensitizers to γ-irradiation in both a nematode in vivo model and breast cancer cells, and could potentially be utilized as an adjuvant therapeutic to enhance radiation sensitivity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Shrivastav M, De Haro LP, Nickoloff JA . Regulation of DNA double-strand break repair pathway choice. Cell Res 2008; 18: 134–147.
Krejci L, Altmannova V, Spirek M, Zhao X . Homologous recombination and its regulation. Nucleic Acids Res 2012; 40: 5795–5818.
Lieber MR . The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79: 181–211.
Zhang C, Wang S, Israel HP, Yan SX, Horowitz DP, Crockford S et al. Higher locoregional recurrence rate for triple-negative breast cancer following neoadjuvant chemotherapy, surgery and radiotherapy. SpringerPlus 2015; 4: 386.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Ambros V . The functions of animal microRNAs. Nature 2004; 431: 350–355.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Adams BD, Kasinski AL, Slack FJ . Aberrant regulation and function of microRNAs in cancer. Curr Biol 2014; 24: R762–R776.
Esquela-Kerscher A, Slack FJ . Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269.
Farazi TA, Spitzer JI, Morozov P, Tuschl T . miRNAs in human cancer. J Pathol 2011; 223: 102–115.
Macfarlane L-A, Murphy PR . MicroRNA: biogenesis, function and role in cancer. Curr Genomics 2010; 11: 537–561.
Garofalo M, Croce CM . MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat 2013; 16: 47–59.
Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015; 518: 107–110.
Peter ME . Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 2010; 29: 2161–2164.
Kai ZS, Pasquinelli AE . MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 2010; 17: 5–10.
Misso G, Teresa M, Martino D, De Rosa G, Farooqi AA, Lombardi A et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 2014; 3: e194.
Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ . Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther 2011; 12: 908–914.
Kato M, Paranjape T, Müller RU, Ullrich R, Nallur S, Gillespie E et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 2009; 28: 2419–2424.
Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 2007; 67: 11111–11116.
Metheetrairut C, Slack FJ . MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev 2013; 23: 12–19.
Weidhaas JB, Eisenmann DM, Holub JM, Nallur SV . A Caenorhabditis elegans tissue model of radiation-induced reproductive cell death. Proc Natl Acad Sci USA 2006; 103: 9946–9951.
Brown JM, Wilson G . Apoptosis genes and resistance to cancer therapy: what does the experimental and clinical data tell us? Cancer Biol Ther 2: 477–490.
Brenner S . The genetics of Caenorhabditis elegans. Genetics 1974; 77: 71–94.
Sulston JE, Horvitz HR . Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 1977; 56: 110–156.
Betel D, Koppal A, Agius P . Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010; 11: R90.
Betel D, Wilson M, Gabow A . The microRNA. org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153.
Friedman R, Farh K . Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105.
Jan C, Friedman R, Ruby J, Bartel D . Formation, regulation and evolution of Caenorhabditis elegans 3 UTRs. Nature 2011; 469: 97–101.
Hammell M, Long D, Zhang L, Lee A . mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein–enriched transcripts. Nat Methods 2008; 5: 813–819.
O'Neil N, Rose A . DNA repair (January 13, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi:10.1895/wormbook.1.54.1, http://www.wormbook.org.
van Haaften G, Romeijn R, Pothof J, Koole W, Mullenders LHF, Pastink A et al. Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Curr Biol 2006; 16: 1344–1350.
Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 2011; 117: 4338–4348.
Feng H, Craig HL, Hope IA . Expression pattern analysis of regulatory transcription factors in Caenorhabditis elegans. Methods Mol Biol 2012; 786: 21–50.
Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL et al. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 let-7 families in the hypodermis and the reproductive system. Dev Dyn 2005; 234: 868–877.
Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S et al. miR-34a silences c-SRC to attenuate tumor growth in triple negative breast cancer. Cancer Res 2015; 76: 927–939.
Zhang Y, Yan L-X, Wu Q-N, Du Z-M, Chen J, Liao D-Z et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res 2011; 71: 3552–3562.
Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou J et al. microRNA-125b inhibits cell migration and invasion by targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett 2013; 5: 829–834.
Tang J, Ahmad A, Sarkar FH . The role of MicroRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci 2012; 13: 13414–13437.
Iliakis G . Cell cycle regulation in irradiated and nonirradiated cells. Semin Oncol 1997; 24: 602–615.
Bernhard EJ, Maity A, Muschel RJ, McKenna WG . Effects of ionizing radiation on cell cycle progression. A review. Radiat Env Biophys 1995; 34: 79–83.
Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff A-K . MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013; 32: 2984–2991.
Mechta-Grigoriou F, Gerald D, Yaniv M . The mammalian Jun proteins: redundancy and specificity. Oncogene 2001; 20: 2378–2389.
Wu L, Fan J, Belasco JG . MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 2006; 103: 4034–4039.
Zhang L, Ge Y, Fuchs E . miR-125b can enhance skin tumor initiation and promote malignant progression by repressing differentiation and prolonging cell survival. Genes Dev 2014; 28: 2532–2546.
Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D . MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 2003; 13: 1317–1323.
Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW et al. The microRNAs of Caenorhabditis elegans. Genes Dev 2003; 17: 991–1008.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 2007; 3: 2395–2403.
Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 2014; 510: 115–120.
Welker NC, Habig JW, Bass BL . Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA 2007; 13: 1090–1102.
Shiiba M, Shinozuka K, Saito K, Fushimi K, Kasamatsu A, Ogawara K et al. MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. Br J Cancer 2013; 108: 1817–1821.
Tan G, Niu J, Shi Y, Ouyang H, Wu ZH . NF-kappaB-dependent microRNA-125b up-regulation promotes cell survival by targeting p38alpha upon ultraviolet radiation. J Biol Chem 2012; 287: 33036–33047.
Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC . Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 2007; 282: 1479–1486.
Rajabi H, Jin C, Ahmad R, McClary AC, Joshi MD, Kufe D . Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer 2010; 1: 62–68.
Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernández-Losa J et al. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One 2013; 8: e76247.
Nyholm AM, Lerche CM, Manfé V, Biskup E, Johansen P, Morling N et al. miR-125b induces cellular senescence in malignant melanoma. BMC Dermatol 2014; 14: 8.
Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer 2011; 128: 1758–1769.
Shi X-B, Xue L, Ma A-H, Tepper CG, Kung H-J, White RW . miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 2011; 71: 538–549.
Gefen N, Binder V, Zaliova M, Linka Y, Morrow M, Novosel A et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 2010; 24: 89–96.
Puissegur MP, Eichner R, Quelen C, Coyaud E, Mari B, Lebrigand K et al. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia 2012; 26: 2224–2232.
Lin K-Y, Ye H, Han B-W, Wang W-T, Wei P-P, He B et al. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene 2015; 35: 3376–3386.
Emmrich S, Rasche M, Schöning J, Reimer C, Keihani S, Maroz A et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev 2014; 28: 858–874.
Zhang L, Stokes N, Polak L, Fuchs E . Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 2011; 8: 294–308.
Ferracin M, Bassi C, Pedriali M, Pagotto S, D’Abundo L, Zagatti B et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol Cancer 2013; 12: 130.
Le MTN, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet 2011; 7: 1–11.
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23: 862–876.
Lowery AJ, Kell MR, Glynn RW, Kerin MJ, Sweeney KJ . Locoregional recurrence after breast cancer surgery: a systematic review by receptor phenotype. Breast Cancer Res Treat 2012; 133: 831–841.
Zaky SS, Lund M, May KA, Godette KD, Beitler JJ, Holmes LR et al. The negative effect of triple-negative breast cancer on outcome after breast-conserving therapy. Ann Surg Oncol 2011; 18: 2858–2865.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007; 13: 4429–4434.
Stiernagle T . Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi:10.1895/wormbook.1.101.1, http://www.wormbook.org.
Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M et al. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res 2007; 35: D472–D475.
Isik M, Berezikov E . Biolistic transformation of Caenorhabditis elegans. Methods Mol Biol 2013; 940: 77–86.
Praitis V, Casey E, Collar D, Austin J . Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 2001; 157: 1217–1226.
Zhao Z, Flibotte S, Murray JI, Blick D, Boyle TJ, Gupta B et al. New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans. Genetics 2010; 184: 853–863.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367.
Olive PL, Banáth JP . The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006; 1: 23–29.
Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR . Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 2008; 9: 9.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001; 25: 402–408.
Acknowledgements
We thank Geoffrey Lyon, Yale Cell Sorter Core Facility, for assistance with fluorescence-activated cell sorting and analysis, the Yale Cesium Irradiator Shared Resource for use of γ-irradiator, Valerie Horsley for use of imaging equipment, and to Shirin Bahmanyar for providing space and making equipment available for these studies. We thank Alan Jiao and Catherine O. Adams for critical reading of this manuscript. Nematode strains used in this work were provided by the C. elegans Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by grants to FJS and JBW from the NIH (R01 CA157749 and R01 CA131301). BDA was supported by a training grant (5T32 HG003198-10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Metheetrairut, C., Adams, B., Nallur, S. et al. cel-mir-237 and its homologue, hsa-miR-125b, modulate the cellular response to ionizing radiation. Oncogene 36, 512–524 (2017). https://doi.org/10.1038/onc.2016.222
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.222
This article is cited by
-
Role of non-coding RNAs in response of breast cancer to radiation therapy
Molecular Biology Reports (2022)
-
Radiation therapy for triple-negative breast cancer: emerging role of microRNAs as biomarkers and radiosensitivity modifiers. A systematic review
Breast Cancer Research and Treatment (2022)
-
Exercise and weight loss interventions and miRNA expression in women with breast cancer
Breast Cancer Research and Treatment (2018)