Abstract
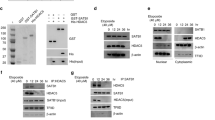
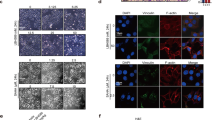
We have previously demonstrated that crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases (HDACs) facilitates breast cancer proliferation. However, the underlying mechanisms are largely unknown. Here, we report that expression of HDAC5 and LSD1 proteins were positively correlated in human breast cancer cell lines and tissue specimens of primary breast tumors. Protein expression of HDAC5 and LSD1 was significantly increased in primary breast cancer specimens in comparison with matched-normal adjacent tissues. Using HDAC5 deletion mutants and co-immunoprecipitation studies, we showed that HDAC5 physically interacted with the LSD1 complex through its domain containing nuclear localization sequence and phosphorylation sites. Although the in vitro acetylation assays revealed that HDAC5 decreased LSD1 protein acetylation, small interfering RNA (siRNA)-mediated HDAC5 knockdown did not alter the acetylation level of LSD1 in MDA-MB-231 cells. Overexpression of HDAC5 stabilized LSD1 protein and decreased the nuclear level of H3K4me1/me2 in MDA-MB-231 cells, whereas loss of HDAC5 by siRNA diminished LSD1 protein stability and demethylation activity. We further demonstrated that HDAC5 promoted the protein stability of USP28, a bona fide deubiquitinase of LSD1. Overexpression of USP28 largely reversed HDAC5-KD-induced LSD1 protein degradation, suggesting a role of HDAC5 as a positive regulator of LSD1 through upregulation of USP28 protein. Depletion of HDAC5 by shRNA hindered cellular proliferation, induced G1 cell cycle arrest, and attenuated migration and colony formation of breast cancer cells. A rescue study showed that increased growth of MDA-MB-231 cells by HDAC5 overexpression was reversed by concurrent LSD1 depletion, indicating that tumor-promoting activity of HDAC5 is an LSD1 dependent function. Moreover, overexpression of HDAC5 accelerated cellular proliferation and promoted acridine mutagen ICR191-induced transformation of MCF10A cells. Taken together, these results suggest that HDAC5 is critical in regulating LSD1 protein stability through post-translational modification, and the HDAC5–LSD1 axis has an important role in promoting breast cancer development and progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Lee MG, Wynder C, Cooch N, Shiekhattar R . An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435.
Huang Y, Marton LJ, Woster PM, Casero RA . Polyamine analogues targeting epigenetic gene regulation. Essays Biochem. 2009; 46: 95–110.
Lee M, Wynder C, Cooch N, Shiekhattar R . An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435.
Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007; 128: 505–518.
Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31: 512–520.
Metzger E, Wissmann M, Yin N, Muller J, Schneider R, Peters A et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437: 436–439.
Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA Jr. . Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids 2012; 42: 887–898.
Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A 2007; 104: 8023–8028.
Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y . Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis 2013; 34: 1196–1207.
Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R . Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 2012; 12: 13.
Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res 2009; 15: 7217–7228.
Nowotarski SL, Pachaiyappan B, Holshouser SL, Kutz CJ, Li Y, Huang Y et al. Structure-activity study for (bis)ureidopropyl- and (bis)thioureidopropyldiamine LSD1 inhibitors with 3-5-3 and 3-6-3 carbon backbone architectures. Bioorg Med Chem 2015; 23: 1601–1612.
Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 2015; 28: 57–69.
Yang XJ, Gregoire S . Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol 2005; 25: 2873–2884.
Martin M, Kettmann R, Dequiedt F . Class IIa histone deacetylases: regulating the regulators. Oncogene 2007; 26: 5450–5467.
McKinsey TA, Zhang CL, Lu J, Olson EN . Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000; 408: 106–111.
Martin M, Kettmann R, Dequiedt F . Class IIa histone deacetylases: conducting development and differentiation. Int J Dev Biol 2009; 53: 291–301.
Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10 AT cells. Breast Cancer Res Treat 2001; 65: 101–110.
Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C et al. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 2013; 5: 224–236.
Han X, Gui B, Xiong C, Zhao L, Liang J, Sun L et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: an antibraking system in neural development. Mol Cell 2014; 55: 482–494.
Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S . Epigenetics in breast cancer: what's new? Breast Cancer Res 2011; 13: 225.
Abukhdeir AM, Park BH . P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med 2008; 10: e19.
Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 2003; 22: 2021–2033.
Lin T, Ponn A, Hu X, Law BK, Lu J . Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010; 29: 4896–4904.
Chen WD, Eshleman JR, Aminoshariae MR, Ma AH, Veloso N, Markowitz SD et al. Cytotoxicity and mutagenicity of frameshift-inducing agent ICR191 in mismatch repair-deficient colon cancer cells. J Natl Cancer Inst 2000; 92: 480–485.
Zientek-Targosz H, Kunnev D, Hawthorn L, Venkov M, Matsui S, Cheney RT et al. Transformation of MCF-10A cells by random mutagenesis with frameshift mutagen ICR191: a model for identifying candidate breast-tumor suppressors. Mol Cancer 2008; 7: 51.
Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res 2010; 16: 3240–3252.
He P, Liang J, Shao T, Guo Y, Hou Y, Li Y . HDAC5 promotes colorectal cancer cell proliferation by up-regulating DLL4 expression. Int J. Clin Exp Med 2015; 8: 6510–6516.
Nagasawa S, Sedukhina AS, Nakagawa Y, Maeda I, Kubota M, Ohnuma S et al. LSD1 overexpression is associated with poor prognosis in basal-like breast cancer, and sensitivity to PARP inhibition. PLoS One 2015; 10: e0118002.
Piao L, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R . SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget 2015; 6: 16939–16950.
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y . Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005; 19: 857–864.
Greco TM, Yu F, Guise AJ, Cristea IM . Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteom 2011; 10: M110.004317.
Diefenbacher ME, Popov N, Blake SM, Schulein-Volk C, Nye E, Spencer-Dene B et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest 2014; 124: 3407–3418.
Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 2007; 9: 765–774.
Zhang D, Zaugg K, Mak TW, Elledge SJ . A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006; 126: 529–542.
Sen N, Kumari R, Singh MI, Das S . HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress. Mol Cell 2013; 52: 406–420.
Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol 2009; 4: 522–526.
Luu TH, Morgan RJ, Leong L, Lim D, McNamara M, Portnow J et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res 2008; 14: 7138–7142.
Modesitt SC, Sill M, Hoffman JS, Bender DP . A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol 2008; 109: 182–186.
Blumenschein GR Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL et al. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs 2008; 26: 81–87.
Shaw PG, Chaerkady R, Wang T, Vasilatos S, Huang Y, Van Houten B et al. Integrated proteomic and metabolic analysis of breast cancer progression. PLoS ONE 2013; 8: e76220.
Varghese F, Bukhari AB, Malhotra R, De A . IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014; 9: e96801.
Ishibashi H, Suzuki T, Suzuki S, Moriya T, Kaneko C, Takizawa T et al. Sex steroid hormone receptors in human thymoma. J Clin Endocrinol Metab 2003; 88: 2309–2317.
Huang Y, Hager ER, Phillips DL, Dunn VR, Hacker A, Frydman B et al. A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin Cancer Res 2003; 9: 2769–2777.
Huang Y, Keen J, Pledgie A, Marton L, Zhu T, Sukumar S et al. Polyamine analogues down-regulate estrogen receptor alpha expression in human breast cancer cells. J Biol Chem 2006; 281: 19055–19063.
Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y . Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res Treat 2014; 146: 99–108.
Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE . Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat 2012; 131: 777–789.
Acknowledgements
This work is supported by US Army Breast Cancer Research Program (W81XWH-14-1-0237 to YH; W81XWH-14-1-0238 to NED), Breast Cancer Research Foundation (to NED and SO) and UPCI Genomics Core Facility supported by NCI P30CA047904.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Cao, C., Vasilatos, S., Bhargava, R. et al. Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression. Oncogene 36, 133–145 (2017). https://doi.org/10.1038/onc.2016.186
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.186
This article is cited by
-
RNA expression of 6 genes from metastatic mucosal gastric cancer serves as the global prognostic marker for gastric cancer with functional validation
British Journal of Cancer (2024)
-
The emerging roles of histone demethylases in cancers
Cancer and Metastasis Reviews (2024)
-
Ubiquitin-specific protease 28: the decipherment of its dual roles in cancer development
Experimental Hematology & Oncology (2023)
-
HDAC5 modulates SATB1 transcriptional activity to promote lung adenocarcinoma
British Journal of Cancer (2023)
-
Glucose-sensitive acetylation of Seryl tRNA synthetase regulates lipid synthesis in breast cancer
Signal Transduction and Targeted Therapy (2021)