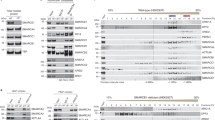
Abstract
As a transcription factor, localization to the nucleus and the recruitment of cofactors to regulate gene transcription is essential. Nuclear localization and nucleosome remodeling and histone deacetylase (NuRD) complex binding are required for the zinc-finger transcription factor CASZ1 to function as a neuroblastoma (NB) tumor suppressor. However, the critical amino acids (AAs) that are required for CASZ1 interaction with NuRD complex and the regulation of CASZ1 subcellular localization have not been characterized. Through alanine scanning, immunofluorescence cell staining and co-immunoprecipitation, we define a critical region at the CASZ1 N terminus (AAs 23–40) that mediates the CASZ1b nuclear localization and NuRD interaction. Furthermore, we identified a nuclear export signal (NES) at the N terminus (AAs 176–192) that contributes to CASZ1 nuclear-cytoplasmic shuttling in a chromosomal maintenance 1-dependent manner. An analysis of CASZ1 protein expression in a primary NB tissue microarray shows that high nuclear CASZ1 staining is detected in tumor samples from NB patients with good prognosis. In contrast, cytoplasmic-restricted CASZ1 staining or low nuclear CASZ1 staining is found in tumor samples from patients with poor prognosis. These findings provide insight into mechanisms by which CASZ1 regulates transcription, and suggests that regulation of CASZ1 subcellular localization may impact its function in normal development and pathologic conditions such as NB tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Accession codes
References
Cui X, Doe CQ . Ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development 1992; 116: 943–952.
Mellerick DM, Kassis JA, Zhang SD, Odenwald WF . Castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron 1992; 9: 789–803.
Charpentier MS, Christine KS, Amin NM, Dorr KM, Kushner EJ, Bautch VL et al. CASZ1 promotes vascular assembly and morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Dev Cell 2013; 25: 132–143.
Liu Z, Li W, Ma X, Ding N, Spallotta F, Southon E et al. Essential role of the zinc finger transcription factor Casz1 for mammalian cardiac morphogenesis and development. J Biol Chem 2014; 289: 29801–29816.
Jordan VK, Zaveri HP, Scott DA . 1p36 deletion syndrome: an update. Appl Clin Genet 2015; 8: 189–200.
Henrich KO, Schwab M, Westermann F . 1p36 tumor suppression—a matter of dosage? Cancer Res 2012; 72: 6079–6088.
Liu Z, Yang X, Li Z, McMahon C, Sizer C, Barenboim-Stapleton L et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ 2011; 18: 1174–1183.
Mattar P, Ericson J, Blackshaw S, Cayouette M . A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 2015; 85: 497–504.
Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation 2010; 121: 2302–2309.
Schmitz M, Driesch C, Beer-Grondke K, Jansen L, Runnebaum IB, Durst M . Loss of gene function as a consequence of human papillomavirus DNA integration. Int J Cancer 2012; 131: E593–E602.
Hoff AM, Johannessen B, Alagaratnam S, Zhao S, Nome T, Lovf M et al. Novel RNA variants in colorectal cancers. Oncotarget 2015; 6: 36587–36602.
Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS et al. EZH2 mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res 2012; 72: 315–324.
Liu Z, Yang X, Tan F, Cullion K, Thiele CJ . Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun 2006; 344: 834–844.
Liu Z, Naranjo A, Thiele CJ . CASZ1b, the short isoform of CASZ1 gene, coexpresses with CASZ1a during neurogenesis and suppresses neuroblastoma cell growth. PLoS One 2011; 6: e18557.
Virden RA, Thiele CJ, Liu Z . Characterization of critical domains within the tumor suppressor CASZ1 required for transcriptional regulation and growth suppression. Mol Cell Biol 2012; 32: 1518–1528.
Liu Z, Lam N, Thiele CJ . Zinc finger transcription factor CASZ1 interacts with histones, DNA repair proteins and recruits NuRD complex to regulate gene transcription. Oncotarget 2015; 6: 27628–27640.
Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst 2009; 101: 663–677.
Schulte JH, Pentek F, Hartmann W, Schramm A, Friedrichs N, Ora I et al. The low-affinity neurotrophin receptor, p75, is upregulated in ganglioneuroblastoma/ganglioneuroma and reduces tumorigenicity of neuroblastoma cells in vivo. Int J Cancer 2009; 124: 2488–2494.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Prieto G, Fullaondo A, Rodriguez JA . Prediction of nuclear export signals using weighted regular expressions (Wregex). Bioinformatics 2014; 30: 1220–1227.
la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S . NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 2003; 31: 393–396.
la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S . Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 2004; 17: 527–536.
Seibel NM, Eljouni J, Nalaskowski MM, Hampe W . Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem 2007; 368: 95–99.
Fornerod M, Ohno M, Yoshida M, Mattaj IW . CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997; 90: 1051–1060.
Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 1999; 96: 9112–9117.
Vacalla CM, Theil T . Cst, a novel mouse gene related to Drosophila Castor, exhibits dynamic expression patterns during neurogenesis and heart development. Mech Dev 2002; 118: 265–268.
Brodeur GM . Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–216.
Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 2009; 27: 289–297.
Maris JM . Recent advances in neuroblastoma. N Engl J Med 2010; 362: 2202–2211.
Rehberg S, Lischka P, Glaser G, Stamminger T, Wegner M, Rosorius O . Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol Cell Biol 2002; 22: 5826–5834.
Kulisz A, Simon HG . An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Mol Cell Biol 2008; 28: 1553–1564.
Malki S, Boizet-Bonhoure B, Poulat F . Shuttling of SOX proteins. Int J Biochem Cell Biol 2010; 42: 411–416.
Rodriguez JA, Henderson BR . Identification of a functional nuclear export sequence in BRCA1. J Biol Chem 2000; 275: 38589–38596.
Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res 2011; 71: 5546–5557.
Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci USA 2007; 104: 20826–20831.
Nie L, Sasaki M, Maki CG . Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J Biol Chem 2007; 282: 14616–14625.
Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM . A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 1999; 18: 1660–1672.
Jiao W, Datta J, Lin HM, Dundr M, Rane SG . Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. J Biol Chem 2006; 281: 38098–38108.
Zhang Y, Xiong Y . A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 2001; 292: 1910–1915.
Liang SH, Clarke MF . A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J Biol Chem 1999; 274: 32699–32703.
Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res 2009; 69: 510–517.
Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012; 120: 1765–1773.
Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood 2011; 118: 3922–3931.
Nguyen KT, Holloway MP, Altura RA . The CRM1 nuclear export protein in normal development and disease. Int J Biochem Mol Biol 2012; 3: 137–151.
Gerecitano J . SINE (selective inhibitor of nuclear export)—translational science in a new class of anti-cancer agents. J Hematol Oncol 2014; 7: 67.
Ishizawa J, Kojima K, Hail N Jr, Tabe Y, Andreeff M . Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther 2015; 153: 25–35.
Denslow SA, Wade PA . The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26: 5433–5438.
Bottardi S, Mavoungou L, Pak H, Daou S, Bourgoin V, Lakehal YA et al. The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet 2014; 10: e1004827.
Krishnakumar R, Kraus WL . PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 2010; 39: 736–749.
Warner LR, Babbitt CC, Primus AE, Severson TF, Haygood R, Wray GA . Functional consequences of genetic variation in primates on tyrosine hydroxylase (TH) expression in vitro. Brain Res 2009; 1288: 1–8.
Winter C, Pawel B, Seiser E, Zhao H, Raabe E, Wang Q et al. Neural cell adhesion molecule (NCAM) isoform expression is associated with neuroblastoma differentiation status. Pediatr Blood Cancer 2008; 51: 10–16.
Acknowledgements
We thank Dr Gregory Wray from the Institute for Genome Sciences and Policy, Duke University for generously providing the tyrosine hydroxylase promoter-pGL4.1-luc (TH-Luc) construct; Nisha Pawer and Neeraj Prasad of the POB, NCI for making the CASZ1b and mutant-EGFP constructs. We appreciate the insightful discussions with Drs Dinah Singer and David Levens of the CCR, NCI on this study. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute and Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Liu, Z., Lam, N., Wang, E. et al. Identification of CASZ1 NES reveals potential mechanisms for loss of CASZ1 tumor suppressor activity in neuroblastoma. Oncogene 36, 97–109 (2017). https://doi.org/10.1038/onc.2016.179
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.179
This article is cited by
-
Role of the CASZ1 transcription factor in tissue development and disease
European Journal of Medical Research (2023)
-
Multidimensional quantitative phenotypic and molecular analysis reveals neomorphic behaviors of p53 missense mutants
npj Breast Cancer (2023)
-
CASZ1: a promising factor modulating aldosterone biosynthesis and mineralocorticoid receptor activity
Hypertension Research (2023)
-
The genetic landscape and clinical implication of pediatric Moyamoya angiopathy in an international cohort
European Journal of Human Genetics (2023)
-
Mutations of 1p genes do not consistently abrogate tumor suppressor functions in 1p-intact neuroblastoma
BMC Cancer (2022)