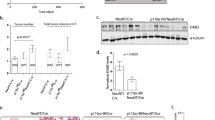
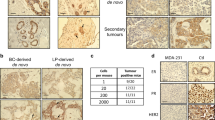
Abstract
Amplification and overexpression of erbB2/neu proto-oncogene is observed in 20–30% human breast cancer and is inversely correlated with the survival of the patient. Despite this, somatic activating mutations within erbB2 in human breast cancers are rare. However, we have previously reported that a splice isoform of erbB2, containing an in-frame deletion of exon 16 (herein referred to as ErbB2ΔEx16), results in oncogenic activation of erbB2 because of constitutive dimerization of the ErbB2 receptor. Here, we demonstrate that the ErbB2ΔEx16 is a major oncogenic driver in breast cancer that constitutively signals from the cell surface. We further show that inducible expression of the ErbB2ΔEx16 variant in mammary gland of transgenic mice results in the rapid development of metastatic multifocal mammary tumors. Genetic and biochemical characterization of the ErbB2ΔEx16-derived mammary tumors exhibit several unique features that distinguish this model from the conventional ErbB2 ones expressing the erbB2 proto-oncogene in mammary epithelium. Unlike the wild-type ErbB2-derived tumors that express luminal keratins, ErbB2ΔEx16-derived tumors exhibit high degree of intratumoral heterogeneity co-expressing both basal and luminal keratins. Consistent with these distinct pathological features, the ErbB2ΔEx16 tumors exhibit distinct signaling and gene expression profiles that correlate with activation of number of key transcription factors implicated in breast cancer metastasis and cancer stem cell renewal.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Andrulis IL, Bull SB, Blackstein ME, Sutherland D, Mak C, Sidlofsky S et al. Neu/erbB-2 amplification identifies a poor-prognosis group of women with node-negative breast cancer. Toronto Breast Cancer Study Group. J Clin Oncol 1998; 16: 1340–1349.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL . Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182.
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE et al. Studies of the HER-2/neu proto-oncogene in human breast and ovaian cancer. Science 1989; 244: 707–712.
Guy CT, Cardiff RD, Muller WJ . Activated neu induces rapid tumor progression. J Biol Chem 1996; 271: 7673–7678.
Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P . Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 1988; 54: 105–115.
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ . Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA 1992; 89: 10578–10582.
Chan R, Muller WJ, Siegel PM . Oncogenic activating mutations in the neu/erbB-2 oncogene are involved in the induction of mammary tumors. Ann N Y Acad Sci 1999; 889: 45–51.
Siegel PM, Dankort DL, Hardy WR, Muller WJ . Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol 1994; 14: 7068–7077.
Siegel PM, Muller WJ . Mutations affecting conserved cysteine residues within the extracellular domain of Neu promote receptor dimerization and activation. Proc Natl Acad Sci USA 1996; 93: 8878–8883.
Kwong KY, Hung MC . A novel splice variant of HER2 with increased transformation activity. Mol Carcinog 1998; 23: 62–68.
Siegel PM, Ryan ED, Cardiff RD, Muller WJ . Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J 1999; 18: 2149–2164.
Alajati A, Sausgruber N, Aceto N, Duss S, Sarret S, Voshol H et al. Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res 2014; 73: 5320–5327.
Castagnoli L, Iezzi M, Ghedini GC, Ciravolo V, Marzano G, Lamolinara A et al. Activated d16HER2 homodimers and Src kinase mediate optimal efficacy for trastuzumab. Cancer Res 2014; 74: 6248–6259.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Ursini-Siegel J, Rajput AB, Lu H, Sanguin-Gendreau V, Zuo D, Papavasiliou V et al. Elevated expression of DecR1 impairs ErbB2/Neu-induced mammary tumor development. Mol Cell Biol 2007; 27: 6361–6371.
Diessner J, Bruttel V, Stein RG, Horn E, Hausler SF, Dietl J et al. Targeting of preexisting and induced breast cancer stem cells with trastuzumab and trastuzumab emtansine (T-DM1). Cell Death Dis 2014; 5: e1149.
Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2002; 2: 451–461.
Finkle D, Quan ZR, Asghari V, Kloss J, Ghaboosi N, Mai E et al. HER2-targeted therapy reduces incidence and progression of midlife mammary tumors in female murine mammary tumor virus huHER2-transgenic mice. Clin Cancer Res 2004; 10: 2499–2511.
DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res 2009; 69: 5364–5373.
Mitra D, Brumlik MJ, Okamgba SU, Zhu Y, Duplessis TT, Parvani JG et al. An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Mol Cancer Ther 2009; 8: 2152–2162.
Clynes RA, Towers TL, Presta LG, Ravetch JV . Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med 2000; 6: 443–446.
Castiglioni F, Tagliabue E, Campiglio M, Pupa SM, Balsari A, Menard S . Role of exon-16-deleted HER2 in breast carcinomas. Endocrine-Related Cancer 2006; 13: 221–232.
McCormack VA, Burton A, Dos-Santos-Silva I, Hipwell JH, Dickens C, Salem D et al. International consortium on mammographic density: methodology and population diversity captured across 22 countries. Cancer Epidemiol 2015; 40: 141–151.
Chen Y, Terajima M, Yang Y, Sun L, Ahn YH, Pankova D et al. Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J Clin Invest 2015; 125: 1147–1162.
Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med 2014; 20: 360–367.
Korsching E, Packeisen J, Liedtke C, Hungermann D, Wulfing P, van Diest PJ et al. The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J Pathol 2005; 206: 451–457.
Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev 2009; 23: 1737–1742.
Ranger JJ, Levy DE, Shahalizadeh S, Hallet M, Muller WJ . Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res 2009; 69: 6823–6830; In Press.
Jones LM, Broz ML, Ranger JJ, Ozcelik J, Ahn R, Zuo D-M et al. Stat3 establishes an immunosuppressive microenvironment during the early stages of breast carcinogenesis to promote tumor growth and metastasis. Cancer Res 2015; 76: 1416–1428.
Andrechek ER, Hardy WR, Girgis-Gabardo AA, Perry RL, Butler R, Graham FL et al. ErbB2 is required for muscle spindle and myoblast cell survival. Mol Cell Biol 2002; 22: 4714–4722.
Perry MC, Dufour CR, Eichner LJ, Tsang DW, Deblois G, Muller WJ et al. ERBB2 deficiency alters an E2F-1-dependent adaptive stress response and leads to cardiac dysfunction. Mol Cell Biol 2014; 34: 4232–4243.
Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A . SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol 2015; 12: 1139–1151.
Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD et al. Alternative expression analysis by RNA sequencing. Nat Methods 2010; 7: 843–847.
Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ . Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res 2009; 69: 5057–5064.
Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci USA 2007; 104: 20302–20307.
Smyth GK, Michaud J, Scott HS . Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics (Oxford, England) 2005; 21: 2067–2075.
Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007; 23: 2700–2707.
Smyth GK, Speed T . Normalization of cDNA microarray data. Methods 2003; 31: 265–273.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014; 10: e1003731.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357.
Chang JT, Carvalho C, Mori S, Bild AH, Gatza ML, Wang Q et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell 2009; 34: 104–114.
Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA 2010; 107: 6994–6999.
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform 2013; 14: 128.
Marcotte R, Zhou L, Kim H, Roskelly CD, Muller WJ . c-Src associates with ErbB2 through an interaction between catalytic domains and confers enhanced transforming potential. Mol Cell Biol 2009; 29: 5858–5871.
Acknowledgements
We thank Dr Jonathan Rayment, Dr Dongmei Zuo, and Ms Virginie Sanguin-Gendreau for their important involvement in this project. This study was supported by grants awarded to WJM from the Terry Fox Foundation (#020002), the Canadian Institutes of Health Research (MOP 93525 and MOP 89751) and the National Institutes of Health PO1 (2PO1CA099031-06A1). WJM is supported by CRC Chair in Molecular Oncology. JT was supported by the Department of Defense Breast Cancer Predoctoral Traineeship award #W81XWH 10-1-0114. ZCH was supported by Susan G Komen Breast Cancer Foundation (CCR14299200) and NIH-NCI (T32-CA009111). ERA is supported by NIH grant R01CA160514. RDC is supported by a US-NCI grant U01 CA141582. MMTV/ErbB2 transgenic mice were a generous donation from Genentech.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Turpin, J., Ling, C., Crosby, E. et al. The ErbB2ΔEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene 35, 6053–6064 (2016). https://doi.org/10.1038/onc.2016.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.129