Abstract
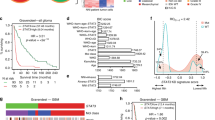
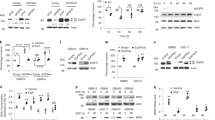
To date, the mutational status of EGFR and PTEN has been shown as relevant for favoring pro- or anti-tumor functions of STAT3 in human glioblastoma multiforme (GBM). We have screened genomic data from 154 patients and have identified a strong positive correlation between STAT3 and HDAC7 expression. In the current work we show the existence of a subpopulation of patients overexpressing HDAC7 and STAT3 that has particularly poor clinical outcome. Surprisingly, the somatic mutation rate of both STAT3 and HDAC7 was insignificant in GBM comparing with EGFR, PTEN or TP53. Depletion of HDAC7 in a range of GBM cells induced the expression of tyrosine kinase JAK1 and the tumor suppressor AKAP12. Both proteins synergistically sustained the activity of STAT3 by inducing its phosphorylation (JAK1) and protein expression (AKAP12). In absence of HDAC7, activated STAT3 was responsible for significant imbalance of secreted pro-/anti-angiogenic factors. This inhibited the migration and sprouting of endothelial cells in paracrine fashion in vitro as well as angiogenesis in vivo. In a murine model of GBM, induced HDAC7-silencing decreased the tumor burden by threefold. The current data show for the first time that silencing HDAC7 can reset the tumor suppressor activity of STAT3, independently of the EGFR/PTEN/TP53 background of the GBM. This effect could be exploited to overcome tumor heterogeneity and provide a new rationale behind the development of specific HDAC7 inhibitors for clinical use.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD . Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res 1990; 50: 6039–6044.
Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA . Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res 2002; 62: 3729–3735.
Talasila KM, Soentgerath A, Euskirchen P, Rosland GV, Wang J, Huszthy PC et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol 2013; 125: 683–698.
Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 2013; 45: 1141–1149.
De Witt Hamer PC . Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol 2010; 12: 304–316.
Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA 2012; 109: 3041–3046.
Inda MM, Bonavia R, Seoane J . Glioblastoma multiforme: a look inside its heterogeneous nature. Cancers 2014; 6: 226–239.
Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 2010; 24: 1731–1745.
Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 2005; 102: 5814–5819.
Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res 2012; 72: 1614–1620.
Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011; 20: 810–817.
Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP . Pten is essential for embryonic development and tumour suppression. Nat Genet 1998; 19: 348–355.
Monk M, Holding C . Human embryonic genes re-expressed in cancer cells. Oncogene 2001; 20: 8085–8091.
Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 1997; 278: 477–483.
de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA et al. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci 2008; 28: 5870–5878.
Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 2013; 24: 438–449.
de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev 2008; 22: 449–462.
Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997; 275: 1943–1947.
Hollander MC, Blumenthal GM, Dennis PA . PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer 2011; 11: 289–301.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Turtoi A, Peixoto P, Castronovo V, Bellahcène A . Histone deacetylases and cancer-associated angiogenesis: current understanding of the biology and clinical perspectives. Crit Rev Oncog 2015; 20: 119–137.
Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN . Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 2006; 126: 321–334.
Mottet D, Bellahcène A, Pirotte S, Waltregny D, Deroanne C, Lamour V et al. Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ Res 2007; 101: 1237–1246.
Turtoi A, Mottet D, Matheus N, Dumont B, Peixoto P, Hennequiere V et al. The angiogenesis suppressor gene AKAP12 is under the epigenetic control of HDAC7 in endothelial cells. Angiogenesis 2012; 15: 543–554.
Bezecny P . Histone deacetylase inhibitors in glioblastoma: pre-clinical and clinical experience. Med Oncol 2014; 31: 985.
Xu WS, Parmigiani RB, Marks PA . Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007; 26: 5541–5552.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE . Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 2005; 307: 269–273.
Xiao H, Chung J, Kao HY, Yang YC . Tip60 is a co-repressor for STAT3. J Biol Chem 2003; 278: 11197–11204.
Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene 2005; 24: 3397–3408.
Wheeler SE, Suzuki S, Thomas SM, Sen M, Leeman-Neill RJ, Chiosea SI et al. Epidermal growth factor receptor variant III mediates head and neck cancer cell invasion via STAT3 activation. Oncogene 2010; 29: 5135–5145.
Schneller D, Machat G, Sousek A, Proell V, van Zijl F, Zulehner G et al. p19(ARF) /p14(ARF) controls oncogenic functions of signal transducer and activator of transcription 3 in hepatocellular carcinoma. Hepatology 2011; 54: 164–172.
Yu H, Lee H, Herrmann A, Buettner R, Jove R . Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014; 14: 736–746.
Atkinson GP, Nozell SE, Benveniste ET . NF-kappa Band STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother 2010; 10: 575–586.
Turtoi A, Blomme A, Castronovo V . Intratumoral heterogeneity and consequences for targeted therapies. Bull Cancer 2015; 102: 17–23.
DiDonato JA, Mercurio F, Karin M . NF-κB and the link between inflammation and cancer. Immunol Rev 2012; 246: 379–400.
Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH et al. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med 2003; 9: 900–906.
Gelman IH . Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev 2012; 31: 493–500.
Su B, Gao L, Meng F, Guo LW, Rothschild J, Gelman IH . Adhesion-mediated cytoskeletal remodeling is controlled by the direct scaffolding of Src from FAK complexes to lipid rafts by SSeCKS/AKAP12. Oncogene 2013; 32: 2016–2026.
Goeppert B, Schmidt CR, Geiselhart L, Dutruel C, Capper D, Renner M et al. Differential expression of the tumor suppressor A-kinase anchor protein 12 in human diffuse and pilocytic astrocytomas is regulated by promoter methylation. J Neuropathol Exp Neurol 2013; 72: 933–941.
West AC, Johnstone RW . New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014; 124: 30–39.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Jaffe EA, Nachman RL, Becker CG, Minick CR . Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973; 52: 2745–2756.
Emi N, Friedmann T, Yee JK . Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol 1991; 65: 1202–1207.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG . Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010; 8: e1000412.
Suresh K . An overview of randomization techniques: an unbiased assessment of outcome in clinical research. J Hum Reprod Sci 2011; 4: 8–11.
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA et al. TM4 microarray software suite. Methods Enzymol 2006; 411: 134–193.
Acknowledgements
The authors acknowledge the experimental support of Dr Chantal Humblet and Ms Alice Marquet (GIGA-histology platform, ULg), Dr Sandra Ormenese (GIGA-imaging platform, ULg) as well as of Evgenia Turtoi (Metastasis Research Laboratory) for the immunohistochemistry analysis and of Naima Maloujahmoum (Metastasis Research Laboratory) for the cell culture. The authors are also thankful to Dr Stephanie Gofflot from the institutional Biobank of the University Hospital Liege for providing GBM fresh tissues and to Dr Gilles Doumont (group of Prof Serge Goldman, Université libre de Bruxelles) for help with animal experiments. The authors are grateful to Ms. Ana Turtoi for proofreading the manuscript. The results shown in this work are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This work was supported with grants from the University of Liège (Concerted Research Action Program (IDEA project)) and from the National Fund for Scientific Research (FNRS/TELEVIE). Andrei Turtoi is a post-doctoral research fellow (FNRS/TELEVIE) and Akeila Bellahcène is a senior research associate (FNRS). Arnaud Blomme is a post-doctoral research fellow (FNRS/TELEVIE). The Center for Microscopy and Molecular Imaging (CMMI) is supported by the European Regional Development Fund and Wallonia. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Peixoto, P., Blomme, A., Costanza, B. et al. HDAC7 inhibition resets STAT3 tumorigenic activity in human glioblastoma independently of EGFR and PTEN: new opportunities for selected targeted therapies. Oncogene 35, 4481–4494 (2016). https://doi.org/10.1038/onc.2015.506
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.506
This article is cited by
-
Bioinformatic identification and experiment validation reveal 6 hub genes, promising diagnostic and therapeutic targets for Alzheimer’s disease
BMC Medical Genomics (2024)
-
AKAP12 promotes cancer stem cell-like phenotypes and activates STAT3 in colorectal cancer
Clinical and Translational Oncology (2023)
-
MiR-489 inhibited the development of gastric cancer via regulating HDAC7 and PI3K/AKT pathway
World Journal of Surgical Oncology (2020)
-
HDAC7 promotes the oncogenicity of nasopharyngeal carcinoma cells by miR-4465-EphA2 signaling axis
Cell Death & Disease (2020)
-
ZNF326 promotes malignant phenotype of glioma by up-regulating HDAC7 expression and activating Wnt pathway
Journal of Experimental & Clinical Cancer Research (2019)