Abstract
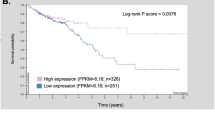
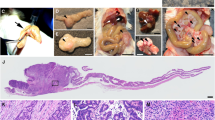
CFTR, the cystic fibrosis (CF) gene, encodes for the CFTR protein that plays an essential role in anion regulation and tissue homeostasis of various epithelia. In the gastrointestinal (GI) tract CFTR promotes chloride and bicarbonate secretion, playing an essential role in ion and acid–base homeostasis. Cftr has been identified as a candidate driver gene for colorectal cancer (CRC) in several Sleeping Beauty DNA transposon-based forward genetic screens in mice. Further, recent epidemiological and clinical studies indicate that CF patients are at high risk for developing tumors in the colon. To investigate the effects of CFTR dysregulation on GI cancer, we generated ApcMin mice that carried an intestinal-specific knockout of Cftr. Our results indicate that Cftr is a tumor suppressor gene in the intestinal tract as Cftr mutant mice developed significantly more tumors in the colon and the entire small intestine. In Apc+/+ mice aged to ~1 year, Cftr deficiency alone caused the development of intestinal tumors in >60% of mice. Colon organoid formation was significantly increased in organoids created from Cftr mutant mice compared with wild-type controls, suggesting a potential role of Cftr in regulating the intestinal stem cell compartment. Microarray data from the Cftr-deficient colon and the small intestine identified dysregulated genes that belong to groups of immune response, ion channel, intestinal stem cell and other growth signaling regulators. These associated clusters of genes were confirmed by pathway analysis using Ingenuity Pathway Analysis and gene set enrichment analysis (GSEA). We also conducted RNA Seq analysis of tumors from Apc+/+ Cftr knockout mice and identified sets of genes dysregulated in tumors including altered Wnt β-catenin target genes. Finally we analyzed expression of CFTR in early stage human CRC patients stratified by risk of recurrence and found that loss of expression of CFTR was significantly associated with poor disease-free survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
De Lisle RC, Borowitz D . The cystic fibrosis intestine. Cold Spring Harb Perspect Med 2013; 3: a009753.
Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989; 245: 1066–1073.
Pedersen SF, Stock C . Ion channels and transporters in cancer: pathophysiology, regulation, and clinical potential. Cancer Res 2013; 73: 1658–1661.
Neglia JP, FitzSimmons SC, Maisonneuve P, Schöni MH, Schöni-Affolter F, Corey M et al. The risk of cancer among patients with cystic fibrosis. Cystic Fibrosis and Cancer Study Group. N Engl J Med 1995; 332: 494–499.
McWilliams R, Highsmith WE, Rabe KG, de Andrade M, Tordsen LA, Holtegaard LM et al. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut 2005; 54: 1661–1662.
Maisonneuve P, Marshall BC, Knapp EA, Lowenfels AB . Cancer risk in cystic fibrosis: a 20-year nationwide study from the United States. J Natl Cancer Inst 2013; 105: 122–129.
Maisonneuve P, FitzSimmons SC, Neglia JP, Campbell PW III, Lowenfels AB . Cancer risk in nontransplanted and transplanted cystic fibrosis patients: a 10-year study. J Natl Cancer Inst 2003; 95: 381–387.
Gelfond D, Borowitz D . Gastrointestinal complications of cystic fibrosis. Clin Gastroenterol Hepatol 2013; 11: 333–342.
Zhang JT, Jiang XH, Xie C, Cheng H, Da Dong J, Wang Y et al. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochim Biophys Acta 2013; 1833: 2961–2969.
Xie C, Jiang XH, Zhang JT, Sun TT, Dong JD, Sanders AJ et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene 2013; 32: 2282–2291, 2291.e1–7.
Sun TT, Wang Y, Cheng H, Xiao HZ, Xiang JJ, Zhang JT et al. Disrupted interaction between CFTR and AF-6/afadin aggravates malignant phenotypes of colon cancer. Biochim Biophys Acta 2014; 1843: 618–628.
Oh SC, Park YY, Park ES, Lim JY, Kim SM, Kim SB et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut 2012; 61: 1291–1298.
Billings JL, Dunitz JM, McAllister S, Herzog T, Bobr A, Khoruts A . Early colon screening of adult patients with cystic fibrosis reveals high incidence of adenomatous colon polyps. J Clin Gastroenterol 2014; 48: e85–e88.
Wilke M, Buijs-Offerman RM, Aarbiou J, Colledge WH, Sheppard DN, Touqui L et al. Mouse models of cystic fibrosis: phenotypic analysis and research applications. J Cyst Fibros 2011; 10 (Suppl 2): S152–S171.
Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323: 1747–1750.
March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet 2011; 43: 1202–1209.
Takeda H, Wei Z, Koso H, Rust AG, Yew CC, Mann MB et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet 2015; 47: 142–150.
Hodges CA, Cotton CU, Palmert MR, Drumm ML . Generation of a conditional null allele for Cftr in mice. Genesis 2008; 46: 546–552.
Snouwaert JN, Brigman KK, Latour AM, Malouf NN, Boucher RC, Smithies O et al. An animal model for cystic fibrosis made by gene targeting. Science 1992; 257: 1083–1088.
Xu Z, Gupta V, Lei D, Holmes A, Carlson E, Gruenert DC . In-frame elimination of exon 10 in Cftrtm1Unc CF mice. Gene 1998; 211: 117–123.
De Lisle RC, Mueller R, Boyd M . Impaired mucosal barrier function in the small intestine of the cystic fibrosis mouse. J Pediatr Gastroenterol Nutr 2011; 53: 371–379.
Than BL, Goos JA, Sarver AL, O'Sullivan MG, Rod A, Starr TK et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 2014; 33: 3861–3868.
de Sousa E, Melo F, Colak S, Buikhuisen J, Koster J, Cameron K et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011; 9: 476–485.
Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011; 8: 511–524.
Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J 2012; 31: 3079–3091.
Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M et al. The intestinal Wnt/TCF signature. Gastroenterology 2007; 132: 628–632.
De Sousa E, Melo F, Wang X, Jansen M, Fessler E, Trinh A et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013; 19: 614–618.
De Lisle RC . Disrupted tight junctions in the small intestine of cystic fibrosis mice. Cell Tissue Res 2014; 355: 131–142.
Clarke LL, Gawenis LR, Bradford EM, Judd LM, Boyle KT, Simpson JE et al. Abnormal Paneth cell granule dissolution and compromised resistance to bacterial colonization in the intestine of CF mice. Am J Physiol Gastrointest Liver Physiol 2004; 286: G1050–G1058.
Norkina O, Kaur S, Ziemer D, De Lisle RC . Inflammation of the cystic fibrosis mouse small intestine. Am J Physiol Gastrointest Liver Physiol 2004; 286: G1032–G1041.
Munck A . Cystic fibrosis: evidence for gut inflammation. Int J Biochem Cell Biol 2014; 52: 180–183.
Juric M, Xiao F, Amasheh S, May O, Wahl K, Bantel H et al. Increased epithelial permeability is the primary cause for bicarbonate loss in inflamed murine colon. Inflamm Bowel Dis 2013; 19: 904–911.
Vij N, Mazur S, Zeitlin PL . CFTR is a negative regulator of NFkappaB mediated innate immune response. PLoS One 2009; 4: e4664.
Chen J, Jiang XH, Chen H, Guo JH, Tsang LL, Yu MK et al. CFTR negatively regulates cyclooxygenase-2-PGE(2) positive feedback loop in inflammation. J Cell Physiol 2012; 227: 2759–2766.
Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and angiogenin-4. PLoS One 2012; 7: e38798.
Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 2013; 342: 447–453.
Schütte A, Ermund A, Becker-Pauly C, Johansson ME, Rodriguez-Pineiro AM, Bäckhed F et al. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci USA 2014; 111: 12396–12401.
Garcia MA, Yang N, Quinton PM . Normal mouse intestinal mucus release requires cystic fibrosis transmembrane regulator-dependent bicarbonate secretion. J Clin Invest 2009; 119: 2613–2622.
Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011; 29: 1120–1127.
Medema JP, Vermeulen L . Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nat Rev 2011; 474: 318–326.
Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015; 47: 312–319.
Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 2013; 19: 619–625.
Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002; 295: 1726–1729.
Yang K, Popova NV, Yang W, Lozonschi I, Tadesse S, Kent S et al. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res 2008; 68: 7313–7322.
Denamur E, Chehab FF . Methylation status of CpG sites in the mouse and human CFTR promoters. DNA Cell Biol 1995; 14: 811–815.
Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology 2011; 16: 1203–1209.
Collawn JF, Fu L, Bartoszewski R, Matalon S . Rescuing ΔF508 CFTR with trimethylangelicin, a dual-acting corrector and potentiator. Am J Physiol Lung Cell Mol Physiol 2014; 307: L431–L434.
Roth EK, Hirtz S, Duerr J, Wenning D, Eichler I, Seydewitz HH et al. The K+ channel opener 1-EBIO potentiates residual function of mutant CFTR in rectal biopsies from cystic fibrosis patients. PLoS One 2011; 6: e24445.
Clarke LL, Gawenis LR, Franklin CL, Harline MC . Increased survival of CFTR knockout mice with an oral osmotic laxative. Lab Anim Sci 1996; 46: 612–618.
Fijneman RJ, Anderson RA, Richards E, Liu J, Tijssen M, Meijer GA et al. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci 2012; 103: 593–599.
Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology 2003; 124: 762–777.
IPA Ingenuity Systems. Available at http://www.ingenuity.com (Accessed 2 February 2013).
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011; 141: 1762–1772.
Acknowledgements
This work was supported by the grant from the National Cancer Institute (NCI) to RC and DL (NCI R01 CA134759-01A1); grant from the Mezin-Koats Colon Cancer Research fund to TS; grant from NCI to support the University of Minnesota Masonic Cancer Center (NCI P30-CA775598); grant from the Whiteside Institute for Clinical Research to RC and PS; grant from the University of Minnesota Foundation to RC, TS and DL; grant from Essentia Health Systems to RC; grant from the AICR (14–1164) to LV; grants from the Dutch Cancer Society (UVA2011–4969, UVA2014–7245) to LV; NWO gravitation grant to JL; grants from the National Cancer Institute to TS (NCI 5R00CA151672-04 and P30-CA77598). We also wish to thank Drs Mitchell Drumm and Craig Hodges of CWRU for providing the Cftr mutant mice.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Than, B., Linnekamp, J., Starr, T. et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene 35, 4191–4199 (2016). https://doi.org/10.1038/onc.2015.483
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.483
This article is cited by
-
SLC26A9 promotes colorectal tumorigenesis by modulating Wnt/β-catenin signaling
Cell Death Discovery (2024)
-
Identification of GUCA2A and COL3A1 as prognostic biomarkers in colorectal cancer by integrating analysis of RNA-Seq data and qRT-PCR validation
Scientific Reports (2023)
-
Loss of Pkd1 limits susceptibility to colitis and colorectal cancer
Oncogenesis (2023)
-
Cystic fibrosis transmembrane conductance regulator (CFTR): beyond cystic fibrosis
Egyptian Journal of Medical Human Genetics (2022)
-
SLC26A9 deficiency causes gastric intraepithelial neoplasia in mice and aggressive gastric cancer in humans
Cellular Oncology (2022)