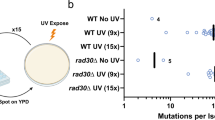
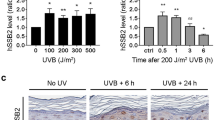
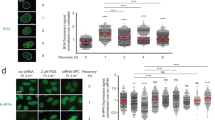
Abstract
The main risk factor for skin cancer is ultraviolet (UV) exposure, which causes DNA damage. Cells respond to UV-induced DNA damage by activating the intra-S-phase checkpoint, which prevents replication fork collapse, late origin firing and stabilizes fragile sites. Recently, the 54-kDa multifunctional protein NONO was found to be involved in the non-homologous end-joining DNA repair process and in poly ADP-ribose polymerase 1 activation. Interestingly, NONO is mutated in several tumour types and emerged as a crucial factor underlying both melanoma development and progression. Therefore, we set out to evaluate whether NONO could be involved in the DNA-damage response to UV radiations. We generated NONO-silenced HeLa cell clones and found that lack of NONO decreased cell growth rate. Then, we challenged NONO-silenced cells with exposure to UV radiations and found that NONO-silenced cells, compared with control cells, continued to synthesize DNA, failed to block new origin firing and impaired CHK1S345 phosphorylation showing a defective checkpoint activation. Consistently, NONO is present at the sites of UV-induced DNA damage where it localizes to RAD9 foci. To position NONO in the DNA-damage response cascade, we analysed the loading onto chromatin of various intra-S-phase checkpoint mediators and found that NONO favours the loading of topoisomerase II-binding protein 1 acting upstream of the ATM and Rad3-related kinase activity. Strikingly, re-expression of NONO, through an sh-resistant mRNA, rescued CHK1S345 phosphorylation in NONO-silenced cells. Interestingly, NONO silencing affected cell response to UV radiations also in a melanoma cell line. Overall, our data uncover a new role for NONO in mediating the cellular response to UV-induced DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Branzei D, Foiani M . The checkpoint response to replication stress. DNA Repair (Amst) 2009; 8: 1038–1046.
Harper JW, Elledge SJ . The DNA damage response: ten years after. Mol Cell 2007; 28: 739–745.
Flynn RL, Zou L . ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 2010; 36: 133–140.
Zou L, Elledge SJ . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300: 1542–1548.
Ellison V, Stillman B . Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol 2003; 1: E33.
Zou L, Liu D, Elledge SJ . Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 2003; 100: 13827–13832.
Kumagai A, Lee J, Yoo HY, Dunphy WG . TopBP1 activates the ATR-ATRIP complex. Cell 2006; 124: 943–955.
Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 2011; 43: 192–202.
Mordes DA, Glick GG, Zhao R, Cortez D . TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 2008; 22: 1478–1489.
Zhao H, Piwnica-Worms H . ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 2001; 21: 4129–4139.
Cimprich KA, Cortez D . ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9: 616–627.
Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 2009; 35: 228–239.
Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ . A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 2012; 14: 318–328.
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316: 1160–1166.
Shav-Tal Y, Zipori D . PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett 2002; 531: 109–114.
Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S et al. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 2005; 25: 6734–6746.
Zhang Z, Carmichael GG . The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001; 106: 465–475.
Straub T, Knudsen BR, Boege F . PSF/p54(nrb) stimulates "jumping" of DNA topoisomerase I between separate DNA helices. Biochemistry 2000; 39: 7552–7558.
Bladen CL, Udayakumar D, Takeda Y, Dynan WS . Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem 2005; 280: 5205–5210.
Li S, Kuhne WW, Kulharya A, Hudson FZ, Ha K, Cao Z et al. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res 2009; 37: 6746–6753.
Rajesh C, Baker DK, Pierce AJ, Pittman DL . The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res 2011; 39: 132–145.
Krietsch J, Caron MC, Gagne JP, Ethier C, Vignard J, Vincent M et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res 2012; 40: 10287–10301.
Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y . Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 2010; 9: 1568–1576.
Kuhnert A, Schmidt U, Monajembashi S, Franke C, Schlott B, Grosse F et al. Proteomic identification of PSF and p54(nrb) as TopBP1-interacting proteins. J Cell Biochem 2012; 113: 1744–1753.
Ishiguro H, Uemura H, Fujinami K, Ikeda N, Ohta S, Kubota Y . 55 kDa nuclear matrix protein (nmt55) mRNA is expressed in human prostate cancer tissue and is associated with the androgen receptor. Int J Cancer 2003; 105: 26–32.
Nelson LD, Bender C, Mannsperger H, Buergy D, Kambakamba P, Mudduluru G et al. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol Cancer 2012; 11: 38.
Pavao M, Huang YH, Hafer LJ, Moreland RB, Traish AM . Immunodetection of nmt55/p54nrb isoforms in human breast cancer. BMC Cancer 2001; 1: 15.
Schiffner S, Zimara N, Schmid R, Bosserhoff AK . p54nrb is a new regulator of progression of malignant melanoma. Carcinogenesis 2011; 32: 1176–1182.
Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997; 15: 2233–2239.
Skalsky YM, Ajuh PM, Parker C, Lamond AI, Goodwin G, Cooper CS . PRCC the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene 2001; 20: 178–187.
Banck MS, Kanwar R, Kulkarni AA, Boora GK, Metge F, Kipp BR et al. The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest 2013; 123: 2502–2508.
Kaufmann WK . The human intra-S checkpoint response to UVC-induced DNA damage. Carcinogenesis 2010; 31: 751–765.
Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 2000; 14: 1448–1459.
Willis N, Rhind N . Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Div 2009; 4: 13.
Zou L, Cortez D, Elledge SJ . Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 2002; 16: 198–208.
Parrilla-Castellar ER, Arlander SJ, Karnitz L . Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004; 3: 1009–1014.
Medhurst AL, Warmerdam DO, Akerman I, Verwayen EH, Kanaar R, Smits VA et al. ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. J Cell Sci 2008; 121: 3933–3940.
Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A et al. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 2001; 411: 969–974.
Oakley GG, Patrick SM . Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci (Landmark Ed) 2010; 15: 883–900.
Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res 2012; 40: 10780–10794.
Stracker TH, Petrini JH . The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 2011; 12: 90–103.
Duursma AM, Driscoll R, Elias JE, Cimprich KA . A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 2013; 50: 116–122.
Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM . The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 2007; 21: 1472–1477.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Dong B, Horowitz DS, Kobayashi R, Krainer AR . Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res 1993; 21: 4085–4092.
Talman V, Tuominen RK, Boije af Gennas G, Yli-Kauhaluoma J, Ekokoski E . C1 Domain-targeted isophthalate derivatives induce cell elongation and cell cycle arrest in HeLa cells. PLoS One 2011; 6: e20053.
Sievers C, Billig G, Gottschalk K, Rudel T . Prohibitins are required for cancer cell proliferation and adhesion. PLoS One 2010; 5: e12735.
Martinez-Ramos I, Maya-Mendoza A, Gariglio P, Aranda-Anzaldo A . A global but stable change in HeLa cell morphology induces reorganization of DNA structural loop domains within the cell nucleus. J Cell Biochem 2005; 96: 79–88.
Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T et al. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci USA 2013; 110: 1592–1599.
Li S, Li Z, Shu FJ, Xiong H, Phillips AC, Dynan WS . Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res 2014; 42: 9771–9780.
Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell 2006; 23: 331–341.
Furuya K, Poitelea M, Guo L, Caspari T, Carr AM . Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev 2004; 18: 1154–1164.
Rappas M, Oliver AW, Pearl LH . Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 2011; 39: 313–324.
Lee J, Kumagai A, Dunphy WG . The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 2007; 282: 28036–28044.
Lee J, Dunphy WG . Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell 2010; 21: 926–935.
Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD et al. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell 2014; 53: 235–246.
Swift LH, Golsteyn RM . Genotoxic anti-cancer agents and their relationship to DNA damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Int J Mol Sci 2014; 15: 3403–3431.
Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J . Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 2006; 26: 6056–6064.
Ma HT, Poon RY . Synchronization of HeLa cells. Methods Mol Biol 2011; 761: 151–161.
Ishii T, Shiomi Y, Takami T, Murakami Y, Ohnishi N, Nishitani H . Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells. J Biol Chem 2010; 285: 41993–42000.
Jackson DA, Pombo A . Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 1998; 140: 1285–1295.
Acknowledgements
We are thankful to the Sbarro Health Research Organization (http://www.shro.org), the Human Health Foundation (http://www.hhfonlus.org), the Commonwealth of Pennsylvania and the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 15690) for their support. We are grateful to Pasquale Barba and Laura Pisapia (Consiglio Nazionale delle Ricerche-Institute of Genetics and Biophysics, Naples, Italy) for the FACS analyses. We also thank Larry M Karnitz, Division of Oncology Research, Mayo Clinic, Rochester, MN, USA for his help in sharing reagents and Marco G Paggi, Regina Elena National Cancer Institute, Rome, Italy for the M14 cell line. Prof Giordano is the director of research line 3 at CROM, Istituto Nazionale Tumori 'Fondazione G. Pascale'- IRCCS, Naples, Italy.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Alfano, L., Costa, C., Caporaso, A. et al. NONO regulates the intra-S-phase checkpoint in response to UV radiation. Oncogene 35, 567–576 (2016). https://doi.org/10.1038/onc.2015.107
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.107
This article is cited by
-
Liquid-liquid phase separation in DNA double-strand breaks repair
Cell Death & Disease (2023)
-
Stabilization of SAMHD1 by NONO is crucial for Ara-C resistance in AML
Cell Death & Disease (2022)
-
Targeting the p300/NONO axis sensitizes melanoma cells to BRAF inhibitors
Oncogene (2021)
-
PRMT1 enhances oncogenic arginine methylation of NONO in colorectal cancer
Oncogene (2021)
-
SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability
Nature Communications (2019)