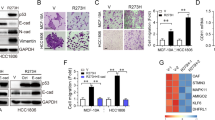
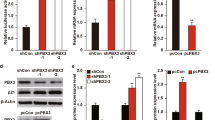
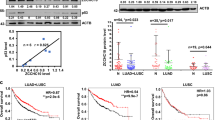
Abstract
Most p53 mutations in human cancers are missense mutations resulting in a full-length mutant p53 protein. Besides losing tumor suppressor activity, some hotspot p53 mutants gain oncogenic functions. This effect is mediated in part, through gene expression changes due to inhibition of p63 and p73 by mutant p53 at their target gene promoters. Here, we report that the tumor suppressor microRNA let-7i is downregulated by mutant p53 in multiple cell lines expressing endogenous mutant p53. In breast cancer patients, significantly decreased let-7i levels were associated with missense mutations in p53. Chromatin immunoprecipitation and promoter luciferase assays established let-7i as a transcriptional target of mutant p53 through p63. Introduction of let-7i to mutant p53 cells significantly inhibited migration, invasion and metastasis by repressing a network of oncogenes including E2F5, LIN28B, MYC and NRAS. Our findings demonstrate that repression of let-7i expression by mutant p53 has a key role in enhancing migration, invasion and metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006; 124: 207–219.
Vousden KH, Lu X . Live or let die: the cell's response to p53. Nat Rev Cancer 2002; 2: 594–604.
Freed-Pastor WA, Prives C . Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268–1286.
Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989; 342: 705–708.
Muller PA, Vousden KH . p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B et al. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 2009; 137: 87–98.
Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 2012; 32: 1252–1265.
Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012; 148: 244–258.
Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 2011; 20: 79–91.
Liu K, Ling S, Lin WC . TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol 2011; 31: 4464–4481.
Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006; 10: 191–202.
Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev 1999; 13: 2490–2501.
Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH et al. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem 2001; 276: 39359–39367.
Li Y, Prives C . Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 2007; 26: 2220–2225.
Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C . A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 2001; 21: 1874–1887.
Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 2000; 25: 47–54.
Di Como CJ, Gaiddon C, Prives C . p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol 1999; 19: 1438–1449.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Djuranovic S, Nahvi A, Green R . miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012; 336: 237–240.
Thomas M, Lieberman J, Lal A . Desperately seeking microRNA targets. Nat Struct Mol Biol 2010; 17: 1169–1174.
Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3'UTR microRNA recognition elements. Mol Cell 2009; 35: 610–625.
Chi SW, Hannon GJ, Darnell RB . An alternative mode of microRNA target recognition. Nat Struct Mol Biol 2012; 19: 321–327.
He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Roush S, Slack FJ . The let-7 family of microRNAs. Trends Cell Biol 2008; 18: 505–516.
Hermeking H . MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 2012; 12: 613–626.
Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007; 26: 745–752.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007; 26: 731–743.
Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 2013; 32: 2992–3000.
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67: 7713–7722.
O'Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, Chen XM et al. NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem 2010; 285: 216–225.
Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev 2012; 26: 830–845.
Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin Jr. WG . Chemosensitivity linked to p73 function. Cancer Cell 2003; 3: 403–410.
Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem 2002; 277: 18817–18826.
Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008; 7: 759–764.
Thomson DW, Bracken CP, Szubert JM, Goodall GJ . On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One 2013; 8: e55214.
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP . The impact of microRNAs on protein output. Nature 2008; 455: 64–71.
Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006; 384: 51–61.
Forman JJ, Legesse-Miller A, Coller HA . A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008; 105: 14879–14884.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67: 9762–9770.
Scian MJ, Stagliano KE, Anderson MA, Hassan S, Bowman M, Miles MF et al. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol Cell Biol 2005; 25: 10097–10110.
Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP . Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol 1998; 18: 3735–3743.
King CE, Wang L, Winograd R, Madison BB, Mongroo PS, Johnstone CN et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 2011; 30: 4185–4193.
Wolfer A, Ramaswamy S . MYC and metastasis. Cancer Res 2011; 71: 2034–2037.
Donzelli S, Fontemaggi G, Fazi F, Di Agostino S, Padula F, Biagioni F et al. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ 2011; 19: 1038–1048.
Wang W, Cheng B, Miao L, Mei Y, Wu M . Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis 2013; 4: e574.
Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene 2012; 32: 3286–3295.
Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell 2010; 17: 273–285.
Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA 2009; 106: 3384–3389.
Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011; 147: 81–94.
Yang WH, Lan HY, Huang CH, Tai SK, Tzeng CH, Kao SY et al. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol 2012; 14: 366–374.
Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood 2011; 118: 903–915.
Hamano R, Miyata H, Yamasaki M, Sugimura K, Tanaka K, Kurokawa Y et al. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer 2012; 106: 1415–1423.
Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet 2011; 7: e1002363.
Prueitt RL, Boersma BJ, Howe TM, Goodman JE, Thomas DD, Ying L et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int Cancer J Int Cancer 2007; 120: 796–805.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Sudha Sharma was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number SC1GM093999. We thank K Prasanth, W Bodmer, B Hassel, M Thomas, J Lieberman and M Gorospe for their comments on this manuscript. We thank B Vogelstein for the DLD1 isogenic cell lines, K Vousden for the H1299-EV and H1299-p53R273H cells, S O’Hara for the pGL4-let-7i and Z Mourelators for the Ago2 (2A8) antibody.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Subramanian, M., Francis, P., Bilke, S. et al. A mutant p53/let-7i-axis-regulated gene network drives cell migration, invasion and metastasis. Oncogene 34, 1094–1104 (2015). https://doi.org/10.1038/onc.2014.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.46
This article is cited by
-
Rewired m6A epitranscriptomic networks link mutant p53 to neoplastic transformation
Nature Communications (2023)
-
CircFAM13B promotes the proliferation of hepatocellular carcinoma by sponging miR-212, upregulating E2F5 expression and activating the P53 pathway
Cancer Cell International (2021)
-
Targeting mutant p53 for cancer therapy: direct and indirect strategies
Journal of Hematology & Oncology (2021)
-
Mutational drivers of cancer cell migration and invasion
British Journal of Cancer (2021)
-
Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes
Nature Communications (2021)