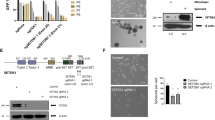
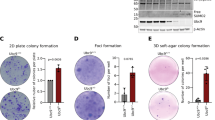
Abstract
Breast cancer is genetically heterogeneous, and recent studies have underlined a prominent contribution of epigenetics to the development of this disease. To uncover new synthetic lethalities with known breast cancer oncogenes, we screened an epigenome-focused short hairpin RNA library on a panel of engineered breast epithelial cell lines. Here we report a selective interaction between the NOTCH1 signaling pathway and the SUMOylation cascade. Knockdown of the E2-conjugating enzyme UBC9 (UBE2I) as well as inhibition of the E1-activating complex SAE1/UBA2 using ginkgolic acid impairs the growth of NOTCH1-activated breast epithelial cells. We show that upon inhibition of SUMOylation NOTCH1-activated cells proceed slower through the cell cycle and ultimately enter apoptosis. Mechanistically, activation of NOTCH1 signaling depletes the pool of unconjugated small ubiquitin-like modifier 1 (SUMO1) and SUMO2/3 leading to increased sensitivity to perturbation of the SUMOylation cascade. Depletion of unconjugated SUMO correlates with sensitivity to inhibition of SUMOylation also in patient-derived breast cancer cell lines with constitutive NOTCH pathway activation. Our investigation suggests that SUMOylation cascade inhibitors should be further explored as targeted treatment for NOTCH-driven breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Maxmen A . The hard facts. Nature 2012; 485: S50–S51.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486: 400–404.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Weinstein IB . Addiction to oncogenes-the Achilles heal of cancer. Science 2002; 297: 63–64.
Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. New Engl J Med 2001; 344: 1031–1037.
Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002; 20: 719–726.
Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New Engl J Med 2010; 363: 809–819.
Luo J, Solimini NL, Elledge SJ . Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009; 136: 823–837.
Kaelin WG . The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 2005; 5: 689–698.
Nijman SMB . Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett 2011; 585: 1–6.
Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci 2001; 98: 10314–10319.
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. New Engl J Med 2009; 361: 123–134.
Mair B, Kubicek S, Nijman SM . Exploiting epigenetic vulnerabilities for cancer therapeutics. Trends Pharmacol Sci 2014; 35: 136–145.
Johnson David G, Dent Sharon YR . Chromatin: receiver and quarterback for cellular signals. Cell 2013; 152: 685–689.
Al-Hussaini H, Subramanyam D, Reedijk M, Sridhar SS . Notch signaling pathway as a therapeutic target in breast cancer. Mol Cancer Ther 2010; 10: 9–15.
Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci 2010; 107: 5012–5017.
Kopan R, Ilagan MXG . The canonical notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233.
Radtke F, Fasnacht N, MacDonald HR . Notch signaling in the immune system. Immunity 2010; 32: 14–27.
Aifantis I, Raetz E, Buonamici S . Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 2008; 8: 380–390.
Ranganathan P, Weaver KL, Capobianco AJ . Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 2011; 11: 338–351.
Grabher C, von Boehmer H, Look AT . Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer 2006; 6: 347–359.
Lobry C, Oh P, Aifantis I . Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med 2011; 208: 1931–1935.
Bray SJ . Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006; 7: 678–689.
Andersson ER, Lendahl U . Therapeutic modulation of Notch signalling—are we there yet? Nat Rev Drug Discov 2014; 13: 357–378.
Falk R, Falk A, Dyson MR, Melidoni AN, Parthiban K, Young JL et al. Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods 2012; 58: 69–78.
Moellering RE, Cornejo M, Davis TN, Bianco CD, Aster JC, Blacklow SC et al. Direct inhibition of the NOTCH transcription factor complex. Nature 2009; 462: 182–188.
Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell Acute lymphoblastic leukemia cells. Cancer Res 2009; 69: 3060–3068.
Purow B . Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol 2012; 727: 305–319.
Robinson DR, Kalyana-Sundaram S, Wu Y-M, Shankar S, Cao X, Ateeq B et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 2011; 17: 1646–1651.
Geiss-Friedlander R, Melchior F . Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8: 947–956.
Gareau JR, Lima CD . The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11: 861–871.
Gill G . SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 2004; 18: 2046–2059.
Seeler J-S, Dejean A . Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 2003; 4: 690–699.
Shiio Y, Eisenman RN . Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci 2003; 100: 13225–13230.
Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 2008; 31: 371–382.
Mo Y-Y, Yu Y, Theodosiou E, Rachel Ee PL, Beck WT . A role for Ubc9 in tumorigenesis. Oncogene 2005; 24: 2677–2683.
Moschos SJ, Smith AP, Mandic M, Athanassiou C, Watson-Hurst K, Jukic DM et al. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: identification of Ubc9 as an important molecule in advanced-stage melanomas. Oncogene 2007; 26: 4216–4225.
Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012; 483: 570–575.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–307.
Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2011; 2: 172–189.
Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol 2011; 7: 787–793.
Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 2009; 16: 133–140.
Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A . Signalling downstream of activated mammalian Notch. Nature 1995; 377: 355–358.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005; 102: 15545–15550.
Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S . SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005; 436: 428–433.
Hay RT . Sumo: a history of modification. Mol Cell 2005; 18: 1–12.
Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 2011; 335: 348–353.
Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET . Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem 1998; 273: 3117–3120.
Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006; 124: 1283–1298.
Sims D, Mendes-Pereira AM, Frankum J, Burgess D, Cerone M-A, Lombardelli C et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol 2011; 12: R104.
Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF . Stem-loop binding protein, the protein that binds the 3' end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 2000; 20: 4188–4198.
Acknowledgements
We thank John Doench, Serena Silver and David E Root (Broad Institute of MIT and Harvard) for shRNA tools and screening protocols; Johannes Bigenzahn (CeMM) for providing the pMSCV-StrepHA-GW-hPGK-Bla vector; and Erika Schirghuber (CeMM) and Berend Snijder (CeMM) for critically reading the manuscript. SK acknowledges support by a Marie Curie Career Integration Grant, the Austrian Federal Ministry of Science, Research and Economy and the National Foundation for Research, Technology and Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Licciardello, M., Müllner, M., Dürnberger, G. et al. NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene 34, 3780–3790 (2015). https://doi.org/10.1038/onc.2014.319
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.319
This article is cited by
-
Transcriptome-wide analysis and modelling of prognostic alternative splicing signatures in invasive breast cancer: a prospective clinical study
Scientific Reports (2020)
-
NICD-mediated notch transduction regulates the different fate of chicken primordial germ cells and spermatogonial stem cells
Cell & Bioscience (2018)
-
Heterozygous IDH1R132H/WT created by “single base editing” inhibits human astroglial cell growth by downregulating YAP
Oncogene (2018)
-
Sumoylation of Notch1 represses its target gene expression during cell stress
Cell Death & Differentiation (2018)
-
SUMO and the robustness of cancer
Nature Reviews Cancer (2017)