Abstract
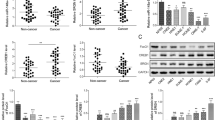
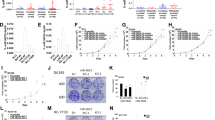
Paired box gene 4 (PAX4) is a transcriptional modulator located on chromosome 7q32, and its expression is dysregulated in a variety of human cancers, suggesting that PAX4 may be important in multiple tumors as a driver gene. Here, we show that PAX4 promoted migration and invasion in human epithelial cancers by decreasing miR-144 and miR-451 (miR-144/451) expression levels. Accordingly, miR-144/451 suppressed the migratory and invasive phenotypes, even in PAX4-expressing cells. Mechanistically, miR-144/451 inhibits cancer metastasis by targeting the A disintegrin and metalloproteinase (ADAM) protein family members ADAMTS5 and ADAM10. Their dysregulation is associated with increased tumor invasiveness and metastasis, then reduced patient prognosis in certain epithelial cancers. This discovery suggests that a PAX4–miR-144/451–ADAMs axis regulates human epithelial cancer metastasis, thus opening up therapeutic possibilities and predicting prognosis for those cancer types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Benfey PN . Molecular biology: microRNA is here to stay. Nature 2003; 425: 244–245.
Vasudevan S, Tong Y, Steitz JA . Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318: 1931–1934.
Guo H, Hu X, Ge S, Qian G, Zhang J . Regulation of RAP1B by miR-139 suppresses human colorectal carcinoma cell proliferation. Int J Biochem Cell Biol 2012; 44: 1465–1472.
Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J . The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer 2013; 12: 77.
Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT . Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 2013; 32: 61–69.
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 2013; 32: 4294–4303.
Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 2010; 207: 1351–1358.
Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis 2012; 33: 2391–2397.
Akiyoshi S, Fukagawa T, Ueo H, Ishibashi M, Takahashi Y, Fabbri M et al. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br J Cancer 2012; 107: 1345–1353.
Tian Y, Nan Y, Han L, Zhang A, Wang G, Jia Z et al. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol 2012; 40: 1105–1112.
Bergamaschi A, Katzenellenbogen BS . Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene 2012; 31: 39–47.
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT et al. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol 2010; 49: 841–850.
Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA 2008; 105: 3333–3338.
Miller DJ, Hayward DC, Reece-Hoyes JS, Scholten I, Catmull J, Gehring WJ et al. Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): implications for the evolution of the Pax gene family. Proc Natl Acad Sci USA 2000; 97: 4475–4480.
Murphy G . The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer 2008; 8: 929–941.
Kozmik Z, Sure U, Ruedi D, Busslinger M, Aguzzi A . Deregulated expression of PAX5 in medulloblastoma. Proc Natl Acad Sci USA 1995; 92: 5709–5713.
Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell 2003; 14: 4376–4386.
Fabbro D, Di Loreto C, Beltrami CA, Belfiore A, Di Lauro R, Damante G . Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res 1994; 54: 4744–4749.
Park SY, Jun JA, Jeong KJ, Heo HJ, Sohn JS, Lee HY et al. Histone deacetylases 1, 6 and 8 are critical for invasion in breast cancer. Oncol Rep 2011; 25: 1677–1681.
Jeon HW, Lee YM . Inhibition of histone deacetylase attenuates hypoxia-induced migration and invasion of cancer cells via the restoration of RECK expression. Mol Cancer Ther 2010; 9: 1361–1370.
Brocker CN, Vasiliou V, Nebert DW . Evolutionary divergence and functions of the ADAM and ADAMTS gene families. Hum Genomics 2009; 4: 43–55.
Lu X, Lu D, Scully M, Kakkar V . ADAM proteins-therapeutic potential in cancer. Curr Cancer Drug Targets 2008; 8: 720–732.
Jones GC . ADAMTS proteinases: potential therapeutic targets? Curr Pharm Biotechnol 2006; 7: 25–31.
Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R et al. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol Ther 2006; 5: 657–664.
Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res 2007; 67: 7703–7712.
Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA 2005; 102: 9182–9187.
Karan D, Lin FC, Bryan M, Ringel J, Moniaux N, Lin MF et al. Expression of ADAMs (a disintegrin and metalloproteases) and TIMP-3 (tissue inhibitor of metalloproteinase-3) in human prostatic adenocarcinomas. Int J Oncol 2003; 23: 1365–1371.
McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM . Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res 2004; 10: 314–323.
Held-Feindt J, Paredes EB, Blomer U, Seidenbecher C, Stark AM, Mehdorn HM et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer 2006; 118: 55–61.
Kumar S, Sharghi-Namini S, Rao N, Ge R . ADAMTS5 functions as an anti-angiogenic and anti-tumorigenic protein independent of its proteoglycanase activity. Am J Pathol 2012; 181: 1056–1068.
Cross NA, Chandrasekharan S, Jokonya N, Fowles A, Hamdy FC, Buttle DJ et al. The expression and regulation of ADAMTS-1, -4, -5, -9, and -15, and TIMP-3 by TGFbeta1 in prostate cells: relevance to the accumulation of versican. Prostate 2005; 63: 269–275.
Cheng C, Li W, Zhang Z, Yoshimura S, Hao Q, Zhang C et al. MicroRNA-144 is regulated by activator protein-1 (AP-1) and decreases expression of Alzheimer disease-related a disintegrin and metalloprotease 10 (ADAM10). J Biol Chem 2013; 288: 13748–13761.
Kobayashi H, Hirata M, Saito T, Itoh S, Chung UI, Kawaguchi H . Transcriptional Induction of ADAMTS5 protein by nuclear factor-kappaB (NF-kappaB) family member RelA/p65 in chondrocytes during osteoarthritis development. J Biol Chem 2013; 288: 28620–28629.
Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum 2009; 60: 2723–2730.
Tamura T, Izumikawa Y, Kishino T, Soejima H, Jinno Y, Niikawa N . Assignment of the human PAX4 gene to chromosome band 7q32 by fluorescence in situ hybridization. Cytogenet Cell Genet 1994; 66: 132–134.
Cigudosa JC, Parsa NZ, Louie DC, Filippa DA, Jhanwar SC, Johansson B et al. Cytogenetic analysis of 363 consecutively ascertained diffuse large B-cell lymphomas. Genes Chromosomes Cancer 1999; 25: 123–133.
Jerkeman M, Johansson B, Akerman M, Cavallin-Stahl E, Kristoffersson U, Mitelman F . Prognostic implications of cytogenetic aberrations in diffuse large B-cell lymphomas. Eur J Haematol 1999; 62: 184–190.
Li Y, Nagai H, Ohno T, Ohashi H, Murohara T, Saito H et al. Aberrant DNA demethylation in promoter region and aberrant expression of mRNA of PAX4 gene in hematologic malignancies. Leuk Res 2006; 30: 1547–1553.
Miyamoto T, Kakizawa T, Ichikawa K, Nishio S, Kajikawa S, Hashizume K . Expression of dominant negative form of PAX4 in human insulinoma. Biochem Biophys Res Commun 2001; 282: 34–40.
Sosa-Pineda B . The gene Pax4 is an essential regulator of pancreatic beta-cell development. Mol Cells 2004; 18: 289–294.
Tokuyama Y, Matsui K, Ishizuka T, Egashira T, Kanatsuka A . The Arg121Trp variant in PAX4 gene is associated with beta-cell dysfunction in Japanese subjects with type 2 diabetes mellitus. Metabolism 2006; 55: 213–216.
Biason-Lauber A, Boehm B, Lang-Muritano M, Gauthier BR, Brun T, Wollheim CB et al. Association of childhood type 1 diabetes mellitus with a variant of PAX4: possible link to beta cell regenerative capacity. Diabetologia 2005; 48: 900–905.
Shikata K, Ninomiya T, Kiyohara Y . Diabetes mellitus and cancer risk: review of the epidemiological evidence. Cancer Sci 2013; 104: 9–14.
Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z . Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 2013; 34: 539–549.
Yan M, Yang X, Wang L, Clark D, Zuo H, Ye D et al. Plasma membrane proteomics of tumor spheres identify CD166 as a novel marker for cancer stem-like cells in head and neck squamous cell carcinoma. Mol Cell Proteomics 2013; 12: 3271–3284.
Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan M et al. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol 2013; 45: 513–520.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 91229103, 81101515, 81201912 and 81272977), the Program of Shanghai Municipal Education Commission (12YZ052 and 12ZZ106) and Project of the Shanghai Science and Technology Committee (11DZ2291800 and 10DZ1951300). We appreciate Liu Jun and Shanghai Cancer Hospital affiliated to Fudan University for providing the 50 pairs of HNSCC tissues. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, J., Qin, X., Sun, Q. et al. Transcriptional control of PAX4-regulated miR-144/451 modulates metastasis by suppressing ADAMs expression. Oncogene 34, 3283–3295 (2015). https://doi.org/10.1038/onc.2014.259
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.259
This article is cited by
-
Global microRNA expression profile in laryngeal carcinoma unveils new prognostic biomarkers and novel insights into field cancerization
Scientific Reports (2022)
-
Iron-sulphur cluster biogenesis factor LYRM4 is a novel prognostic biomarker associated with immune infiltrates in hepatocellular carcinoma
Cancer Cell International (2021)
-
MicroRNA-144 represses gliomas progression and elevates susceptibility to Temozolomide by targeting CAV2 and FGF7
Scientific Reports (2020)
-
LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus
Journal of Experimental & Clinical Cancer Research (2019)
-
miR-144/451 in hematopoiesis and beyond
ExRNA (2019)