Abstract
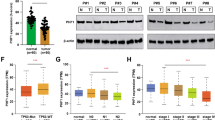
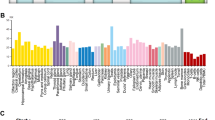
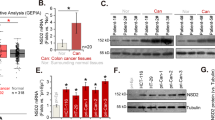
Plant homeodomain finger 2 (PHF2) has a role in epigenetic regulation of gene expression by demethylating H3K9-Me2. Several genome-wide studies have demonstrated that the chromosomal region including the PHF2 gene is often deleted in some cancers including colorectal cancer, and this finding encouraged us to investigate the tumor suppressive role of PHF2. As p53 is a critical tumor suppressor in colon cancer, we tested the possibility that PHF2 is an epigenetic regulator of p53. PHF2 was associated with p53, and thereby, promoted p53-driven gene expression in cancer cells under genotoxic stress. PHF2 converted the chromatin that is favorable for transcription by demethylating the repressive H3K9-Me2 mark. In an HCT116 xenograft model, PHF2 was found to be required for the anticancer effects of oxaliplatin and doxorubicin. In PHF2-deficient xenografts, p53 expression was profoundly induced by both drugs, but its downstream product p21 was not, suggesting that p53 cannot be activated in the absence of PHF2. To find clinical evidence about the role of PHF2, we analyzed the expressions of PHF2, p53 and p21 in human colon cancer tissues and adjacent normal tissues from patients. PHF2 was downregulated in cancer tissues and PHF2 correlated with p21 in cancers expressing functional p53. Colon and stomach cancer tissue arrays showed a positive correlation between PHF2 and p21 expressions. Informatics analyses using the Oncomine database also supported our notion that PHF2 is downregulated in colon and stomach cancers. On the basis of these findings, we propose that PHF2 acts as a tumor suppressor in association with p53 in cancer development and ensures p53-mediated cell death in response to chemotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Levine AJ, Oren M . The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Murray-Zmijewski F, Slee EA, Lu X . A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 2008; 9: 702–712.
Gu W, Shi X-L, Roeder RG . Synergistic activation of transcription by CBP and p53. Nature 1997; 387: 819–823.
Gu W, Roeder RG . Activation of p53 sequence-specific dna binding by acetylation of the p53 C-terminal domain. Cell 1997; 90: 595–606.
Sakaguchi K, Herrera JE, Si Saito, Miki T, Bustin M, Vassilev A et al. DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev 1998; 12: 2831–2841.
Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M et al. Role of the Histone Acetyl Transferase Tip60 in the p53 Pathway. J Biol Chem 2004; 279: 44825–44833.
Jin L, Hanigan CL, Wu Y, Wang W, Park BH, Woster PM et al. Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53- and DNMT1(DNA methyltransferase 1)-independent manner. Biochem J 2013; 449: 459–468.
Wiesner GL, Daley D, Lewis S, Ticknor C, Platzer P, Lutterbaugh J et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci 2003; 100: 12961–12965.
Sinha S, Singh R, Alam N, Roy A, Roychoudhury S, Panda C . Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: Pathological significance in early- and late-onset breast carcinoma. Mol Cancer 2008; 7: 84.
Ghosh A, Ghosh S, Maiti GP, Mukherjee S, Mukherjee N, Chakraborty J et al. Association of FANCC and PTCH1 with the development of early dysplastic lesions of the head and neck. Ann Surg Oncol 2012; 19: 528–538.
Hasenpusch-Theil K, Chadwick BP, Theil T, Heath SK, Wilkinson DG, Frischauf A-M . PHF2 a novel PHD finger gene located on human Chromosome 9q22. Mamm Genome 1999; 10: 294–298.
Wen H, Li J, Song T, Lu M, Kan P-Y, Lee MG et al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem 2010; 285: 9322–9326.
Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol 2011; 13: 668–675.
Okuno Y, Ohtake F, Igarashi K, Kanno J, Matsumoto T, Takada I et al. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes 2012; 33: 1426–1434.
Stender Joshua D, Pascual G, Liu W, Kaikkonen Minna U, Do K, Spann Nathanael J et al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell 2012; 48: 28–38.
Kurdistani SK, Grunstein M . Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 2003; 4: 276–284.
Jaskelioff M, Peterson CL . Chromatin and transcription: histones continue to make their marks. Nat Cell Biol 2003; 5: 395–399.
Schneider R, Grosschedl R . Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 2007; 21: 3027–3043.
Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA . p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol 2012; 19: 478–484.
Fortschegger K, Shiekhattar R . Plant homeodomain fingers form a helping hand for transcription. Epigenetics 2011; 6: 4–8.
Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I . PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol 2010; 17: 445–450.
Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 2010; 38: 165–178.
Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res 2010; 20: 908–918.
Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010; 466: 508–512.
Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS et al. Regulation of p53 activity through lysine methylation. Nature 2004; 432: 353–360.
Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444: 629–632.
Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW et al. Modulation of p53 Function by SET8-Mediated Methylation at Lysine 382. Mol Cell 2007; 27: 636–646.
Munro AJ, Lain S, Lane DP . P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer 2005; 92: 434–444.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Zirbes TK, Baldus SE, Moenig SP, Nolden S, Kunze D, Shafizadeh ST et al. Prognostic impact of p21/waf1/cip1 in colorectal cancer. Int J Cancer 2000; 89: 14–18.
Ogino S, Nosho K, Shima K, Baba Y, Irahara N, Kirkner GJ et al. p21 Expression in Colon Cancer and Modifying Effects of Patient Age and Body Mass Index on Prognosis. Cancer Epidemiol Biomarkers Prev 2009; 18: 2513–2521.
Yeo E-J, Chun Y-S, Cho Y-S, Kim J, Lee J-C, Kim M-S et al. YC-1: A Potential Anticancer Drug Targeting Hypoxia-Inducible Factor 1. J Natl Cancer Ins 2003; 95: 516–525.
Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D et al. Modeling Oncogenic Signaling in Colon Tumors by Multidirectional Analyses of Microarray Data Directed for Maximization of Analytical Reliability. PLoS ONE 2010; 5: e13091.
Acknowledgements
This work was supported by a Korean Health Technology R&D grant (A121106, A120476), National Research Foundation grants of Korea (2009-0066985, 2011-0008634 and 2011-0030737) and by a grant from the Seoul National University Hospital Research Fund 2011 (0320110180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, KH., Park, JW., Sung, HS. et al. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene 34, 2897–2909 (2015). https://doi.org/10.1038/onc.2014.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.219
This article is cited by
-
The emerging roles of histone demethylases in cancers
Cancer and Metastasis Reviews (2024)
-
Phosphorylation of PHF2 by AMPK releases the repressive H3K9me2 and inhibits cancer metastasis
Signal Transduction and Targeted Therapy (2023)
-
Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma
Nature Communications (2023)
-
HIF-1α-mediated augmentation of miRNA-18b-5p facilitates proliferation and metastasis in osteosarcoma through attenuation PHF2
Scientific Reports (2022)
-
A novel role of tumor suppressor ZMYND8 in inducing differentiation of breast cancer cells through its dual-histone binding function
Journal of Biosciences (2020)