Abstract
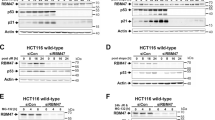
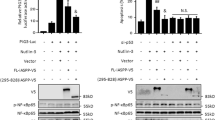
Chromatin conformation has a major role in all cellular decisions. We showed previously that P53 pro-apoptotic target promoters are enriched with H3K9me3 mark and induction of P53 abrogates this repressive chromatin conformation by downregulating SUV39H1, the writer of this mark present on these promoters. In the present study, we demonstrate that in response to P53 stabilization, its pro-apoptotic target promoters become enriched with the H3K4me3 epigenetic mark as well as its readers, Wdr5, RbBP5 and Ash2L, which were not observed in response to SUV39H1 downregulation alone. Overexpression of Ash2L enhanced P53-dependent apoptosis in response to chemotherapy, associated with increased P53 pro-apoptotic gene promoter occupancy and target gene expression. In contrast, pre-silencing of Ash2L abrogated P53’s ability to induce the expression of these transcriptional targets, without affecting P53 or RNAP II recruitment. However, Ash2L pre-silencing, under the same conditions, resulted in reduced RNAP II ser5-CTD phosphorylation on these same pro-apoptotic target promoters, which correlated with reduced promoter occupancy of TFIIB as well as TFIIF (RAP74). Based on these findings, we propose that Ash2L acts in concert with P53 promoter occupancy to activate RNAP II by aiding formation of a stable transcription pre-initiation complex required for its activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jenuwein T, Allis CD . Translating the histone code. Science 2001; 293: 1074–1080.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837.
Schmid CD, Bucher P . ChIP-Seq data reveal nucleosome architecture of human promoters. Cell 2007; 131: 831–832.
Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599.
Beckerman R, Prives C . Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2010; 2: a000935.
Laptenko O, Prives C . Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ 2006; 13: 951–961.
Mandinova A, Lee SW . The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci Transl Med 2011; 3: 64rv1.
Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA . p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol 2012; 19: 478–484.
Lee J, Kim DH, Lee S, Yang QH, Lee DK, Lee SK et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci USA 2009; 106: 8513–8518.
el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Shilatifard A . The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012; 81: 65–95.
Vermeulen M, Timmers HT . Grasping trimethylation of histone H3 at lysine 4. Epigenomics 2010; 2: 395–406.
Bieging KT, Attardi LD . Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol 2012; 22: 97–106.
Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005; 120: 169–181.
Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 2002; 99: 8695–8700.
Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA et al. A high-resolution map of active promoters in the human genome. Nature 2005; 436: 876–880.
Roh TY, Cuddapah S, Zhao K . Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 2005; 19: 542–552.
Roh TY, Cuddapah S, Cui K, Zhao K . The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA 2006; 103: 15782–15787.
Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 2011; 30: 4198–4210.
Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 2013; 154: 297–310.
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 2006; 13: 713–719.
Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J . Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol cell 2006; 22: 137–144.
Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A . Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 2006; 13: 852–854.
Trievel RC, Shilatifard A . WDR5, a complexed protein. Nat Struct Mol Biol 2009; 16: 678–680.
Patel A, Dharmarajan V, Vought VE, Cosgrove MS . On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem 2009; 284: 24242–24256.
Ng HH, Robert F, Young RA, Struhl K . Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 2003; 11: 709–719.
Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR . Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell 2009; 33: 181–191.
Pommier Y, Leo E, Zhang H, Marchand C . DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 2010; 17: 421–433.
Fu Y, Li S, Zu Y, Yang G, Yang Z, Luo M et al. Medicinal chemistry of paclitaxel and its analogues. Curr Med Chem 2009; 16: 3966–3985.
Egloff S, Dienstbier M, Murphy S . Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet 2012; 28: 333–341.
Gomes NP, Espinosa JM . Differential regulation of p53 target genes: it's (core promoter) elementary. Genes Dev 2010; 24: 111–114.
Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ 3rd et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011; 332: 99–103.
Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ . Emerging views on the CTD code. Genet Res Int 2012; 2012: 347214.
Egloff S, Murphy S . Cracking the RNA polymerase II CTD code. Trends Genet 2008; 24: 280–288.
Bartkowiak B, Greenleaf AL . Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription 2011; 2: 115–119.
Thomas MC, Chiang CM . The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41: 105–178.
Shandilya J, Wang Y, Roberts SG . TFIIB dephosphorylation links transcription inhibition with the p53-dependent DNA damage response. Proc Natl Acad Sci USA 2012; 109: 18797–18802.
Zheng H, Chen L, Pledger WJ, Fang J, Chen J . p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene 2014; 6: 734–744.
Bosch-Presegue L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell 2011; 42: 210–223.
Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T . Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 2011; 30: 2817–2828.
Buratowski S . The CTD code. Nat Struct Biol 2003; 10: 679–680.
Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013; 152: 1021–1036.
Deng W, Roberts SG . TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 2007; 116: 417–429.
Wang Y, Fairley JA, Roberts SG . Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol 2010; 20: 548–553.
Cabart P, Ujvari A, Pal M, Luse DS . Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. Proc Natl Acad Sci USA 2011; 108: 15786–15791.
Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K . Recruitment of p300/CBP in p53-dependent signal pathways. Cell 1997; 89: 1175–1184.
Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM . Binding and modulation of p53 by p300/CBP coactivators. Nature 1997; 387: 823–827.
Thomas A, White E . Suppression of the p300-dependent mdm2 negative-feedback loop induces the p53 apoptotic function. Genes Dev 1998; 12: 1975–1985.
Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell 1998; 2: 405–415.
Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK . CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J 2010; 29: 3660–3672.
Goodman RH, Smolik S . CBP/p300 in cell growth, transformation, and development. Genes Dev 2000; 14: 1553–1577.
Levine AJ, Oren M . The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758.
Vousden KH, Prives C . Blinded by the Light: The Growing Complexity of p53. Cell 2009; 137: 413–431.
Murray-Zmijewski F, Slee EA, Lu X . A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 2008; 9: 702–712.
Jaskelioff M, Peterson CL . Chromatin and transcription: histones continue to make their marks. Nat Cell Biol 2003; 5: 395–399.
Grewal SI, Elgin SC . Transcription and RNA interference in the formation of heterochromatin. Nature 2007; 447: 399–406.
Hosoda H, Miyao T, Uchida S, Sakai S, Kida S . Development of a tightly-regulated tetracycline-dependent transcriptional activator and repressor co-expression system for the strong induction of transgene expression. Cytotechnology 2011; 63: 211–216.
Sugrue MM, Shin DY, Lee SW, Aaronson SA . Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci USA 1997; 94: 9648–9653.
Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA 2008; 105: 3933–3938.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848.
Mungamuri SK, Yang X, Thor AD, Somasundaram K . Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res 2006; 66: 4715–4724.
Acknowledgements
We thank SW Lee (MGH, Harvard) for helpful discussions. This research was supported by grants from the National Cancer Institute (P01CA080058) and Breast Cancer Research Foundation (SAA). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Mungamuri, S., Wang, S., Manfredi, J. et al. Ash2L enables P53-dependent apoptosis by favoring stable transcription pre-initiation complex formation on its pro-apoptotic target promoters. Oncogene 34, 2461–2470 (2015). https://doi.org/10.1038/onc.2014.198
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.198
This article is cited by
-
Placental mesenchymal stem cells restore glucose and energy homeostasis in obesogenic adipocytes
Cell and Tissue Research (2023)
-
Epigenetic-focused CRISPR/Cas9 screen identifies (absent, small, or homeotic)2-like protein (ASH2L) as a regulator of glioblastoma cell survival
Cell Communication and Signaling (2023)
-
Human placental mesenchymal stromal cell therapy restores the cytokine efflux and insulin signaling in the skeletal muscle of obesity-induced type 2 diabetes rat model
Human Cell (2022)
-
Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin
Nature Communications (2021)
-
The promise(s) of mesenchymal stem cell therapy in averting preclinical diabetes: lessons from in vivo and in vitro model systems
Scientific Reports (2021)