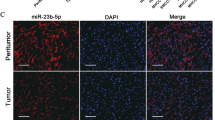
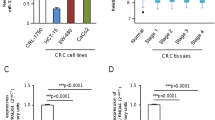
Abstract
It is well known that microRNAs (miRs) are abnormally expressed in various cancers and target the messenger RNAs (mRNAs) of cancer-associated genes. While (miRs) are abnormally expressed in various cancers, whether miRs directly target oncogenic proteins is unknown. The present study investigated the inhibitory effects of miR-18a on colon cancer progression, which was considered to be mediated through its direct binding and degradation of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). An MTT assay and xenograft model demonstrated that the transfection of miR-18a induced apoptosis in SW620 cells. A binding assay revealed direct binding between miR-18a and hnRNP A1 in the cytoplasm of SW620 cells, which inhibited the oncogenic functions of hnRNP A1. A competitor RNA, which included the complementary sequence of the region of the miR-18a-hnRNP A1 binding site, repressed the effects of miR-18a on the induction of cancer cell apoptosis. In vitro single and in vivo double isotope assays demonstrated that miR-18a induced the degradation of hnRNP A1. An immunocytochemical study of hnRNP A1 and LC3-II and the inhibition of autophagy by 3-methyladenine and ATG7, p62 and BAG3 siRNA showed that miR-18a and hnRNP A1 formed a complex that was degraded through the autophagolysosomal pathway. This is the first report showing a novel function of a miR in the autophagolysosomal degradation of an oncogenic protein resulting from the creation of a complex consisting of the miR and a RNA-binding protein, which suppressed cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Capon DJ, Seeburg PH, McGrath JP, Hayflick JS, Edman U, Levinson AD et al. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature 1983; 304: 507–513.
Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M . Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature 1987; 327: 298–303.
Guan RJ, Fu Y, Holt PR, Pardee AB . Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology 1999; 116: 1063–1071.
Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989; 342: 705–708.
Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN et al. APC mutations occur early during colorectal tumorigenesis. Nature 1992; 359: 235–237.
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS . Non-coding RNAs: regulators of disease. J Pathol 2010; 220: 126–139.
Mattick JS, Gagen MJ . The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 2001; 18: 1611–1630.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S . Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11: 228–234.
Eulalio A, Huntzinger E, Izaurralde E . Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132: 9–14.
Filipowicz W, Bhattacharyya SN, Sonenberg N . Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E et al. MicroRNA expression in zebrafish embryonic development. Science 2005; 309: 310–311.
Wightman B, Ha I, Ruvkun G . Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75: 855–862.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435: 828–833.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132: 875–886.
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67: 9762–9770.
Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 2009; 136: 683–693.
Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR et al. Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer. Breast Cancer Res Treat 2011; 127: 69–80.
Tao J, Wu D, Li P, Xu B, Lu Q, Zhang W . microRNA-18a, a member of the oncogenic miR-17-92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol Med Rep 2012; 5: 167–172.
Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 2009; 34: 1069–1075.
Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010; 140: 652–665.
Guil S, Caceres JF . The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 2007; 14: 591–596.
Michlewski G, Guil S, Semple CA, Caceres JF . Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Molecular cell 2008; 32: 383–393.
Ushigome M, Ubagai T, Fukuda H, Tsuchiya N, Sugimura T, Takatsuka J et al. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int J Oncol 2005; 26: 635–640.
Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V . RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ Res 2004; 95: 1058–1066.
Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J . Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem 2008; 283: 23274–23287.
Kirkin V, McEwan DG, Novak I, Dikic I . A role for ubiquitin in selective autophagy. Molecular cell 2009; 34: 259–269.
Kettern N, Rogon C, Limmer A, Schild H, Hohfeld J . The Hsc/Hsp70 co-chaperone network controls antigen aggregation and presentation during maturation of professional antigen presenting cells. PLoS One 2011; 6: e16398.
Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA 2010; 107: 6334–6339.
Tsuchiya N, Izumiya M, Ogata-Kawata H, Okamoto K, Fujiwara Y, Nakai M et al. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res 2011; 71: 4628–4639.
Zhang Y, Wang Z, Chen M, Peng L, Wang X, Ma Q et al. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer 2012; 11: 23.
Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis 2012; 33: 519–528.
Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis 2012; 33: 68–76.
Hope NR, Murray GI . The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum Pathol 2011; 42: 393–402.
Qased AB, Yi H, Liang N, Ma S, Qiao S, Liu X . MicroRNA-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells. Mol Med Report 2013; 7 (2): 559–564.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Fujiya, M., Konishi, H., Mohamed Kamel, M. et al. microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene 33, 4847–4856 (2014). https://doi.org/10.1038/onc.2013.429
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.429
Keywords
This article is cited by
-
Heterogeneous nuclear ribonucleoprotein A/B: an emerging group of cancer biomarkers and therapeutic targets
Cell Death Discovery (2022)
-
RETRACTED ARTICLE: HnRNP A1 - mediated alternative splicing of CCDC50 contributes to cancer progression of clear cell renal cell carcinoma via ZNF395
Journal of Experimental & Clinical Cancer Research (2020)
-
Autophagy in cancers including brain tumors: role of MicroRNAs
Cell Communication and Signaling (2020)
-
A tumor-specific modulation of heterogeneous ribonucleoprotein A0 promotes excessive mitosis and growth in colorectal cancer cells
Cell Death & Disease (2020)
-
MicroRNAs and their role for T stage determination and lymph node metastasis in early colon carcinoma
Clinical & Experimental Metastasis (2017)