Abstract
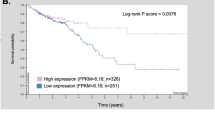
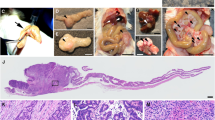
Kcnq1, which encodes for the pore-forming α-subunit of a voltage-gated potassium channel, was identified as a gastrointestinal (GI) tract cancer susceptibility gene in multiple Sleeping Beauty DNA transposon-based forward genetic screens in mice. To confirm that Kcnq1 has a functional role in GI tract cancer, we created ApcMin mice that carried a targeted deletion mutation in Kcnq1. Results demonstrated that Kcnq1 is a tumor suppressor gene as Kcnq1 mutant mice developed significantly more intestinal tumors, especially in the proximal small intestine and colon, and some of these tumors progressed to become aggressive adenocarcinomas. Gross tissue abnormalities were also observed in the rectum, pancreas and stomach. Colon organoid formation was significantly increased in organoids created from Kcnq1 mutant mice compared with wild-type littermate controls, suggesting a role for Kcnq1 in the regulation of the intestinal crypt stem cell compartment. To identify gene expression changes due to loss of Kcnq1, we carried out microarray studies in the colon and proximal small intestine. We identified altered genes involved in innate immune responses, goblet and Paneth cell function, ion channels, intestinal stem cells, epidermal growth factor receptor and other growth regulatory signaling pathways. We also found genes implicated in inflammation and in cellular detoxification. Pathway analysis using Ingenuity Pathway Analysis and Gene Set Enrichment Analysis confirmed the importance of these gene clusters and further identified significant overlap with genes regulated by MUC2 and CFTR, two important regulators of intestinal homeostasis. To investigate the role of KCNQ1 in human colorectal cancer (CRC), we measured protein levels of KCNQ1 by immunohistochemistry in tissue microarrays containing samples from CRC patients with liver metastases who had undergone hepatic resection. Results showed that low expression of KCNQ1 expression was significantly associated with poor overall survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Peroz D, Rodriguez N, Choveau F, Baro I, Merot J, Loussouarn G . Kv7.1 (KCNQ1) properties and channelopathies. J Physiol 2008; 586: 1785–1789.
Winbo A, Sandstrom O, Palmqvist R, Rydberg A . Iron-deficiency anaemia, gastric hyperplasia, and elevated gastrin levels due to potassium channel dysfunction in the Jervell and Lange–Nielsen Syndrome. Cardiol Young 2012; 18: 1–10.
Grahammer F, Herling AW, Lang HJ, Schmitt-Graff A, Wittekindt OH, Nitschke R et al. The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology 2001; 120: 1363–1371.
Rice KS, Dickson G, Lane M, Crawford J, Chung SK, Rees MI et al. Elevated serum gastrin levels in Jervell and Lange–Nielsen syndrome: a marker of severe KCNQ1 dysfunction? Heart Rhythm 2011; 8: 551–554.
Nikou GC, Toubanakis C, Moulakakis KG, Pavlatos S, Kosmidis C, Mallas E et al. Carcinoid tumors of the duodenum and the ampulla of Vater: current diagnostic and therapeutic approach in a series of 8 patients. Case series. Int J Surg 2011; 9: 248–253.
Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008; 40: 1098–1102.
Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008; 40: 1092–1097.
Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323: 1747–1750.
March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet 2011; 43: 1202–1209.
Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA 2012; 109: 5934–5941.
Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012; 486: 266–270.
Starr TK, Scott PM, Marsh BM, Zhao L, Than BL, O'Sullivan MG et al. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proc Natl Acad Sci USA 2011; 108: 5765–5770.
Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD et al. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest 2000; 106: 1447–1455.
Casimiro MC, Knollmann BC, Ebert SN, Vary JC Jr, Greene AE, Franz MR et al. Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange–Nielsen Syndrome. Proc Natl Acad Sci USA 2001; 98: 2526–2531.
Casimiro MC, Knollmann BC, Yamoah EN, Nie L, Vary JC Jr, Sirenko SG et al. Targeted point mutagenesis of mouse Kcnq1: phenotypic analysis of mice with point mutations that cause Romano–Ward syndrome in humans. Genomics 2004; 84: 555–564.
Takagi T, Nishio H, Yagi T, Kuwahara M, Tsubone H, Tanigawa N et al. Phenotypic analysis of vertigo 2 Jackson mice with a Kcnq1 potassium channel mutation. Exp Anim 2007; 56: 295–300.
Elso CM, Lu X, Culiat CT, Rutledge JC, Cacheiro NL, Generoso WM et al. Heightened susceptibility to chronic gastritis, hyperplasia and metaplasia inKcnq1 mutant mice. Hum Mol Genet 2004; 13: 2813–2821.
Demolombe S, Franco D, de Boer P, Kuperschmidt S, Roden D, Pereon Y et al. Differential expression of KvLQT1 and its regulator IsK in mouse epithelia. Am J Physiol Cell Physiol 2001; 280: C359–C372.
Dedek K, Waldegger S . Colocalization of KCNQ1/KCNE channel subunits in the mouse gastrointestinal tract. Pflugers Arch 2001; 442: 896–902.
Vallon V, Grahammer F, Volkl H, Sandu CD, Richter K, Rexhepaj R et al. KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proc Natl Acad Sci USA 2005; 102: 17864–17869.
Warth R, Garcia Alzamora M, Kim JK, Zdebik A, Nitschke R, Bleich M et al. The role of KCNQ1/KCNE1 K(+) channels in intestine and pancreas: lessons from the KCNE1 knockout mouse. Pflugers Arch 2002; 443: 822–828.
Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J 2012; 31: 3079–3091.
IPA Ingenuity Systems. Available at http://www.ingenuity.com (accessed 2 February 2013).
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Norkina O, Kaur S, Ziemer D, De Lisle RC . Inflammation of the cystic fibrosis mouse small intestine. Am J Physiol Gastrointest Liver Physiol 2004; 286: G1032–G1041.
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB . The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7: 11–20.
Knobloch M, Braun SM, Zurkirchen L, von Schoultz C, Zamboni N, Araúzo-Bravo MJ et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 2013; 493: 226–230.
Duerkop BA, Vaishnava S, Hooper LV . Immune responses to the microbiota at the intestinal mucosal surface. Immunity 2009; 31: 368–376.
Shalapour S, Deiser K, Sercan O, Tuckermann J, Minnich K, Willimsky G et al. Commensal microflora and interferon-gamma promote steady-state interleukin-7 production in vivo. Eur J Immunol 2010; 40: 2391–2400.
Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011; 334: 255–258.
Dziarski R, Gupta D . Review: mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun 2010; 16: 168–174.
Kang S, Okuno T, Takegahara N, Takamatsu H, Nojima S, Kimura T et al. Intestinal epithelial cell-derived semaphorin 7A negatively regulates development of colitis via αvβ1 integrin. J Immunol 2012; 188: 1108–1116.
Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012; 487: 477–481.
Lang F, Shumilina E . Regulation of ion channels by the serum- and glucocorticoid-inducible kinase SGK1. FASEB J 2013; 27: 3–12.
De Lisle RC, Mueller R, Boyd M . Impaired mucosal barrier function in the small intestine of the cystic fibrosis mouse. J Pediatr Gastroenterol Nutr 2011; 53: 371–379.
Yang K, Popova NV, Yang W, Lozonschi I, Tadesse S, Kent S et al. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res 2008; 68: 7313–7322.
Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and angiogenin-4. PLoS One 2012; 7: e38798.
Xiong Q, Gao Z, Wang W, Li M . Activation of Kv7 (KCNQ) voltage-gated potassium channels by synthetic compounds. Trends Pharmacol Sci 2008; 29: 99–107.
Wulff H, Castle NA, Pardo LA . Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discovery 2009; 8: 982–1001.
Preston P, Wartosch L, Günzel D, Fromm M, Kongsuphol P, Ousingsawat J et al. Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl− transport. J Biol Chem 2010; 285: 7165–7175.
Maisonneuve P, FitzSimmons SC, Neglia JP, Campbell PW III, Lowenfels AB . Cancer risk in nontransplanted and transplanted cystic fibrosis patients: a 10-year study. J Natl Cancer Inst. 2003; 95: 381–387.
Fijneman RJA, Anderson R, Richards E, Liu J, Tijssen M, Meijer GA et al. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci 2012; 103: 593–92011.
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology Nov 2011; 141: 1762–17772.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Stichting FMWV Rotterdam: Code Goed Gebruik van lichaamsmateriaal 2011, ISBN 978-90-817510-0-1.
Simon R, Mirlacher M, Sauter G . Tissue microarrays. Biotechniques 2004; 36: 98–105.
Belt EJT, Fijneman RJA, van den Berg EG, Bril H, Delis-van Diemen PM, Tijssen M et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J Cancer 2011; 47: 1837–1845.
Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A . Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol 2007; 60: 1112–1116.
Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH . Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg 1999; 230: 309–318.
Acknowledgements
We thank Dr Karl Pfeifer at the NIH for generously providing Kcnq1 mutant mice and reviewing the manuscript. Research was supported by a grant to RC and DL (NCI R01 CA134759-01A1) and to JG and RF (Center for Translational Molecular Medicine, DeCoDe project grant 03O-101) and to TKS (NCI R00 4R00CA151672-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Than, B., Goos, J., Sarver, A. et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 33, 3861–3868 (2014). https://doi.org/10.1038/onc.2013.350
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.350
Keywords
This article is cited by
-
Detection of hypokalemia disorder and its relation with hypercalcemia in blood serum using LIBS technique for patients of colorectal cancer grade I and grade II
Lasers in Medical Science (2022)
-
Age-dependent transition from islet insulin hypersecretion to hyposecretion in mice with the long QT-syndrome loss-of-function mutation Kcnq1-A340V
Scientific Reports (2021)
-
Pathophysiological role of ion channels and transporters in gastrointestinal mucosal diseases
Cellular and Molecular Life Sciences (2021)
-
Selective expression of KCNA5 and KCNB1 genes in gastric and colorectal carcinoma
BMC Cancer (2020)
-
A novel role for estrogen-induced signaling in the colorectal cancer gender bias
Irish Journal of Medical Science (1971 -) (2019)