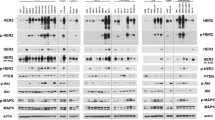
Abstract
Overexpression of the receptor tyrosine kinase ERBB2 (also known as HER2) occurs in around 15% of breast cancers and is driven by amplification of the ERBB2 gene. ERBB2 amplification is a marker of poor prognosis, and although anti-ERBB2-targeted therapies have shown significant clinical benefit, de novo and acquired resistance remains an important problem. Genomic profiling has demonstrated that ERBB2+ve breast cancers are distinguished from ER+ve and ‘triple-negative’ breast cancers by harbouring not only the ERBB2 amplification on 17q12, but also a number of co-amplified genes on 17q12 and amplification events on other chromosomes. Some of these genes may have important roles in influencing clinical outcome, and could represent genetic dependencies in ERBB2+ve cancers and therefore potential therapeutic targets. Here, we describe an integrated genomic, gene expression and functional analysis to determine whether the genes present within amplicons are critical for the survival of ERBB2+ve breast tumour cells. We show that only a fraction of the ERBB2-amplified breast tumour lines are truly addicted to the ERBB2 oncogene at the mRNA level and display a heterogeneous set of additional genetic dependencies. These include an addiction to the transcription factor gene TFAP2C when it is amplified and overexpressed, suggesting that TFAP2C represents a genetic dependency in some ERBB2+ve breast cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344: 783–792.
Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med 2006; 355: 2733–2743.
Vogel C, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L et al. First-line, single-agent Herceptin(R) (trastuzumab) in metastatic breast cancer. A preliminary report. Eur J Cancer 2001; 37 (Suppl 1): 25–29.
Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Davidson NE et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005; 353: 1673–1684.
Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet 2007; 369: 29–36.
Gianni L, Dafni U, Gelber RD, Azambuja E, Muehlbauer S, Goldhirsch A et al. Treatment with trastuzumab for 1 year after adjuvant chemotherapy in patients with HER2-positive early breast cancer: a 4-year follow-up of a randomised controlled trial. Lancet Oncol 2011; 12: 236–244.
Perez EA, Romond EH, Suman VJ, Jeong JH, Davidson NE, Geyer CE et al. Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSABP B-31. J Clin Oncol 2011; 29: 3366–3373.
Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006; 10: 529–541.
Arriola E, Marchio C, Tan DS, Drury SC, Lambros MB, Natrajan R et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest 2008; 88: 491–503.
Staaf J, Jonsson G, Ringner M, Vallon-Christersson J, Grabau D, Arason A et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res 2010; 12: R25.
Shiu KK, Natrajan R, Geyer FC, Ashworth A, Reis-Filho JS . DNA amplifications in breast cancer: genotypic-phenotypic correlations. Future Oncol 2010; 6: 967–984.
Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS . A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 2010; 10: 59–64.
Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res 2009; 15: 2711–2722.
Mackay A, Tamber N, Fenwick K, Iravani M, Grigoriadis A, Dexter T et al. A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res Treat 2009; 118: 481–498.
Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Goransson H, Liljedahl U et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia—a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer 2008; 47: 697–711.
Cleator SJ, Powles TJ, Dexter T, Fulford L, Mackay A, Smith IE et al. The effect of the stromal component of breast tumours on prediction of clinical outcome using gene expression microarray analysis. Breast Cancer Res 2006; 8: R32.
Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 2006; 16: 1465–1479.
Sims D, Bursteinas B, Gao Q, Jain E, MacKay A, Mitsopoulos C et al. ROCK: a breast cancer functional genomics resource. Breast Cancer Res Treat 2010; 124: 567–572.
Sircoulomb F, Bekhouche I, Finetti P, Adelaide J, Ben Hamida A, Bonansea J et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer 2010; 10: 539.
Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DS et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat 2010; 121: 575–589.
Tan DS, Reis-Filho JS . Comparative genomic hybridisation arrays: high-throughput tools to determine targeted therapy in breast cancer. Pathobiology 2008; 75: 63–74.
Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res 2007; 9: R65.
Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010; 29: 2013–2023.
Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS . A census of amplified and overexpressed human cancer genes. Nat Rev 2009; 10: 59–64.
Kao J, Pollack JR . RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 2006; 45: 761–769.
Boutros M, Bras LP, Huber W . Analysis of cell-based RNAi screens. Genome Biol 2006; 7: R66.
Zhang JH, Chung TD, Oldenburg KR . A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73.
Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res 2008; 68: 5878–5887.
Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci Signal 2009; 2: ra31.
Bosher JM, Williams T, Hurst HC . The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA 1995; 92: 744–747.
Vernimmen D, Begon D, Salvador C, Gofflot S, Grooteclaes M, Winkler R . Identification of HTF (HER2 transcription factor) as an AP-2 (activator protein-2) transcription factor and contribution of the HTF binding site to ERBB2 gene overexpression. Biochem J 2003; 370 (Pt 1): 323–329.
Ailan H, Xiangwen X, Daolong R, Lu G, Xiaofeng D, Xi Q et al. Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC Cancer 2009; 9: 279.
Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene 2007; 26: 1959–1970.
Turner BC, Zhang J, Gumbs AA, Maher MG, Kaplan L, Carter D et al. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res 1998; 58: 5466–5472.
Zhao C, Yasui K, Lee CJ, Kurioka H, Hosokawa Y, Oka T et al. Elevated expression levels of NCOA3, TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer 2003; 98: 18–23.
Gee JM, Eloranta JJ, Ibbitt JC, Robertson JF, Ellis IO, Williams T et al. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol 2009; 217: 32–41.
Guler G, Iliopoulos D, Guler N, Himmetoglu C, Hayran M, Huebner K . Wwox and Ap2gamma expression levels predict tamoxifen response. Clin Cancer Res 2007; 13: 6115–6121.
Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ . Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer 2010; 49: 948–962.
Williams CM, Scibetta AG, Friedrich JK, Canosa M, Berlato C, Moss CH et al. AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J 2009; 28: 3591–3601.
Delacroix L, Begon D, Chatel G, Jackers P, Winkler R . Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol 2005; 24: 582–594.
Weinstein IB . Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 2002; 297: 63–64.
Scaltriti M, Eichhorn PJ, Cortes J, Prudkin L, Aura C, Jimenez J et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci USA 2011; 108: 3761–3766.
O’Brien NA, Browne BC, Chow L, Wang Y, Ginther C, Arboleda J et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther 2010; 9: 1489–1502.
Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst 2007; 99: 628–638.
Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res 2008; 68: 9221–9230.
Harris LN, You F, Schnitt SJ, Witkiewicz A, Lu X, Sgroi D et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin Cancer Res 2007; 13: 1198–1207.
Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol 2011; 29: 166–173.
Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, Miles D et al. Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol 2010; 28: 1138–1144.
Marchio C, Natrajan R, Shiu KK, Lambros MB, Rodriguez-Pinilla SM, Tan DS et al. The genomic profile of HER2-amplified breast cancers: the influence of ER status. J Pathol 2008; 216: 399–407.
Pietersen AM, Horlings HM, Hauptmann M, Langerod A, Ajouaou A, Cornelissen-Steijger P et al. EZH2 and BMI1 inversely correlate with prognosis and TP53 mutation in breast cancer. Breast Cancer Res 2008; 10: R109.
Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat 2010; 121: 53–64.
Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I et al. Functional viability profiles of breast cancer. Cancer Discov 2011; 1: 260–273.
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434: 917–921.
Lambros MB, Simpson PT, Jones C, Natrajan R, Westbury C, Steele D et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest 2006; 86: 398–408.
Acknowledgements
This work was supported by grants from Cancer Research UK, AACR as part of the Stand Up To Cancer Breast Cancer Dream Team Initiative, and Breakthrough Breast Cancer. Dr Kai-Keen Shiu is an Avon Clinical Fellow.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Shiu, KK., Wetterskog, D., Mackay, A. et al. Integrative molecular and functional profiling of ERBB2-amplified breast cancers identifies new genetic dependencies. Oncogene 33, 619–631 (2014). https://doi.org/10.1038/onc.2012.625
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.625
Keywords
This article is cited by
-
Co-expression of transcription factor AP-2beta (TFAP2B) and GATA3 in human mammary epithelial cells with intense, apicobasal immunoreactivity for CK8/18
Journal of Molecular Histology (2021)
-
Lobular carcinoma in situ and invasive lobular breast cancer are characterized by enhanced expression of transcription factor AP-2β
Laboratory Investigation (2018)
-
Pathobiological implications of the d16HER2 splice variant for stemness and aggressiveness of HER2-positive breast cancer
Oncogene (2017)
-
A HER2-specific Modified Fc Fragment (Fcab) Induces Antitumor Effects Through Degradation of HER2 and Apoptosis
Molecular Therapy (2015)
-
The role of Tcfap2c in tumorigenesis and cancer growth in an activated Neu model of mammary carcinogenesis
Oncogene (2015)