Abstract
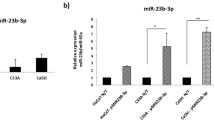
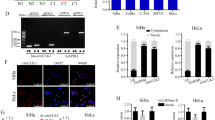
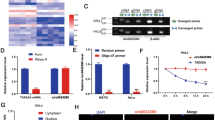
MicroRNAs (miRNAs) act as important gene regulators in human genomes and their aberrant expression links to many malignancies. We previously identified a different characteristic miRNA expression profile in cervical cancer from that in cervical normal tissues, including the downregulated miR-424. However, the role and mechanism of miR-424 in cervical cancer still remain unknown. Here, we focused on identifying the tumor-suppressive function and clinical significance of miR-424 and exploring the mechanistic relevance by characterizing its target. We showed a significantly decreased expression of miR-424 in 147 cervical cancer tissues versus 74 cervical normal tissues by performing quantitative RT–PCR. In 147 cervical cancer tissue samples, low-level expression of miR-424 was positively correlated with poor tumor differentiation, advanced clinical stage, lymph node metastasis and other poor prognostic clinicopathological parameters. Further in vitro observations showed that enforced expression of miR-424 inhibited cell growth by both enhancing apoptosis and blocking G1/S transition, and suppressed cell migration and invasion in two human cervical cancer cell lines, SiHa and CaSki, implying that miR-424 functions as a tumor suppressor in the progression of cervical cancer. Interestingly, overexpression of miR-424 inhibited the expression of protein checkpoint kinase 1 (Chk1) and phosphorylated Chk1 (p-Chk1) at residues Ser345 and decreased the activity of luciferase-reporter containing the 3′-untranslated region (UTR) of Chk1 with predicted miR-424-binding site. Moreover, miR-424 expression levels were inversely correlated with Chk1 and p-Chk1 protein levels in both cervical cancer and normal tissues. Furthermore, RNAi-mediated knockdown of Chk1 decreased matrix metalloproteinase 9 expression and phenocopied the tumor suppressive effects of miR-424 in cell models. Taken together, our results identify a crucial tumor suppressive role of miR-424 in the progression of cervical cancer at least partly via upreglating the expression of Chk1 and p-Chk1, and suggest that miR-424 might be a candidate of prognostic predictor or an anticancer therapeutic target for cervical cancer patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Pillai RS, Bhattacharyya SN, Filipowicz W . Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 2007; 17: 118–126.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Garzon R, Calin GA, Croce CM . MicroRNAs in Cancer. Annu Rev Med 2009; 60: 167–179.
Kent OA, Mendell JT . A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 2006; 25: 6188–6196.
Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH . Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008; 283: 1026–1033.
Selcuklu SD, Donoghue MT, Spillane C . miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 2009; 37 (Part 4): 918–925.
Medina PP, Nolde M, Slack FJ . OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010; 467: 86–90.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14: 2348–2360.
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol 2011; 224: 484–495.
Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA 2007; 104: 19849–19854.
Sarkar S, Dey BK, Dutta A . MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell 2010; 21: 2138–2149.
Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest 2010; 120: 4141–4154.
Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 2008; 36: 5391–5404.
Pallasch CP, Patz M, Park YJ, Hagist S, Eggle D, Claus R et al. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 2009; 114: 3255–3264.
Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res 2011; 39: 1880–1893.
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem 2011; 286: 10725–10734.
Li J, Huang H, Sun L, Yang M, Pan C, Chen W et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res 2009; 15: 3998–4008.
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 2008; 3: e2557.
Witten D, Tibshirani R, Gu SG, Fire A, Lui WO . Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol 2010; 8: 58.
Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA 2011; 108: 3336–3341.
Deryugina EI, Quigley JP . Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev 2006; 25: 9–34.
Yu W, Liu J, Xiong X, Ai Y, Wang H . Expression of MMP9 and CD147 in invasive squamous cell carcinoma of the uterine cervix and their implication. Pathol Res Pract 2009; 205: 709–715.
Lin LF, Chuang CH, Li CF, Liao CC, Cheng CP, Cheng TL et al. ZBRK1 acts as a metastatic suppressor by directly regulating MMP9 in cervical cancer. Cancer Res 2010; 70: 192–201.
Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2010; 24: 460–466.
Kawahigashi Y, Mishima T, Mizuguchi Y, Arima Y, Yokomuro S, Kanda T et al. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J Nihon Med Sch 2009; 76: 188–197.
Rentoft M, Fahlen J, Coates PJ, Laurell G, Sjostrom B, Ryden P et al. miRNA analysis of formalin-fixed squamous cell carcinomas of the tongue is affected by age of the samples. Int J Oncol 2011; 38: 61–69.
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ . Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis 2010; 11: 50–54.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T . Identification of novel genes coding for small expressed RNAs. Science 2001; 294: 853–858.
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ . miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006; 34 (Database issue): D140–D144.
Wang Z, Wang M, Kar S, Carr BI . Involvement of ATM-mediated Chk1/2 and JNK kinase signaling activation in HKH40A-induced cell growth inhibition. J Cell Physiol 2009; 221: 213–220.
Lopez-Girona A, Tanaka K, Chen XB, Baber BA, McGowan CH, Russell P . Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc Natl Acad Sci USA 2001; 98: 11289–11294.
Bartek J, Lukas J . Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 2001; 13: 738–747.
Zhao H, Watkins JL, Piwnica-Worms H . Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA 2002; 99: 14795–14800.
Zachos G, Rainey MD, Gillespie DA . Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 2003; 22: 713–723.
Madoz-Gurpide J, Canamero M, Sanchez L, Solano J, Alfonso P, Casal JI . A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics 2007; 6: 2150–2164.
Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res 2007; 67: 6574–6581.
Ehlen O, Nodin B, Rexhepaj E, Brandstedt J, Uhlen M, Alvarado-Kristensson M et al. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl Oncol 2011; 4: 212–221.
Gao Q, Huang X, Tang D, Cao Y, Chen G, Lu Y et al. Influence of chk1 and plk1 silencing on radiation- or cisplatin-induced cytotoxicity in human malignant cells. Apoptosis 2006; 11: 1789–1800.
Li YL, Ye F, Hu Y, Lu WG, Xie X . Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal Biochem 2009; 394: 110–116.
Shen Y, Li Y, Ye F, Wang F, Lu W, Xie X . Identification of suitable reference genes for measurement of gene expression in human cervical tissues. Anal Biochem 2010; 405: 224–229.
Li YL, Ye F, Cheng XD, Hu Y, Zhou CY, Lu WG et al. Identification of glia maturation factor beta as an independent prognostic predictor for serous ovarian cancer. Eur J Cancer 2010; 46: 2104–2118.
Ye F, Hu Y, Lu W, Zhou C, Xie X . Expression of leukaemia inhibitory factor in epithelial ovarian carcinoma: correlation with clinical characteristics. Histopathology 2008; 53: 224–228.
Acknowledgements
We thank pathologist Xiaoduan Chen for histological diagnoses of cervical tissues and Dr Yifan Cheng for sample collections. We also thank continuous financial support by grants from the National Natural Science Foundation of China (Grant No. 81172475), Zhejiang Provincial Natural Science Foundation of China (Grant No. Z2110056), Zhejiang Provincial Medical and Health Science and Technology Project (Grant No. 2009A132 and No. 2011ZDA015) and Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Disclaimer
The authors declare that the material is an original research, has not been previously published and has not been submitted for publication elsewhere while under consideration.
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Xu, J., Li, Y., Wang, F. et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 32, 976–987 (2013). https://doi.org/10.1038/onc.2012.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.121
Keywords
This article is cited by
-
Role of miR-424 in the carcinogenesis
Clinical and Translational Oncology (2023)
-
Suppressive effects of umbilical cord mesenchymal stem cell-derived exosomal miR-15a-5p on the progression of cholangiocarcinoma by inhibiting CHEK1 expression
Cell Death Discovery (2022)
-
DSCAM-AS1 promotes cervical carcinoma cell proliferation and invasion via sponging miR-338-3p
Environmental Science and Pollution Research (2022)
-
MiR-139-3p Targets CHEK1 Modulating DNA Repair and Cell Viability in Lung Squamous Carcinoma Cells
Molecular Biotechnology (2022)
-
LINC00922 promotes the proliferation, migration, invasion and EMT process of liver cancer cells by regulating miR-424-5p/ARK5
Molecular and Cellular Biochemistry (2021)