Abstract
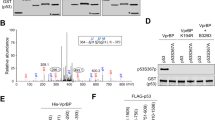
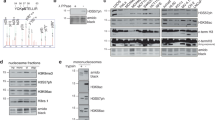
Linker histone H1.2 has been shown to suppress p53-dependent transcription through the modulation of chromatin remodeling; however, little is known about the mechanisms governing the antagonistic effects of H1.2 in DNA damage response. Here, we show that the repressive action of H1.2 on p53 function is negatively regulated via acetylation of p53 C-terminal regulatory domain and phosphorylation of H1.2 C-terminal tail. p53 acetylation by p300 impairs the interaction of p53 with H1.2 and triggers a rapid activation of p53-dependent transcription. Similarly, DNA-PK-mediated phosphorylation of H1.2 at T146 enhances p53 transcriptional activity by impeding H1.2 binding to p53 and thereby attenuating its suppressive effects on p53 transactivation. Consistent with these findings, point mutations mimicking modification states of H1.2 and p53 lead to a significant increase in p53-induced apoptosis. These data suggest that p53 acetylation–H1.2 phosphorylation cascade serves as a unique mechanism for triggering p53-dependent DNA damage response pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
An W, Kim J, Roeder RG . (2004). Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748.
Ausio J . (2006). Histone variants--the structure behind the function. Brief Funct Genomic Proteomic 5: 228–243.
Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD et al. (2001). Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254.
Beckerman R, Prives C . (2010). Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935.
Brown DT . (2003). Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 81: 221–227.
Bustin M, Catez F, Lim JH . (2005). The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620.
Caterino TL, Hayes JJ . (2011). Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol 89: 35–44.
Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE . (2003). The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol 23: 8626–8636.
Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA . (1999). Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol Cell 4: 641–647.
Espinosa JM, Verdun RE, Emerson BM . (2003). p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027.
Georgel PT, Hansen JC . (2001). Linker histone function in chromatin: dual mechanisms of action. Biochem Cell Biol 79: 313–316.
Gu W, Roeder RG . (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606.
Hale TK, Contreras A, Morrison AJ, Herrera RE . (2006). Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol Cell 22: 693–699.
Happel N, Doenecke D . (2009). Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431: 1–12.
Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA et al. (2002). Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9: 263–267.
Izzo A, Kamieniarz K, Schneider R . (2008). The histone H1 family: specific members, specific functions? Biol Chem 389: 333–343.
Jaskelioff M, Gavin IM, Peterson CL, Logie C . (2000). SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol 20: 3058–3068.
Junttila MR, Evan GI . (2009). p53--a Jack of all trades but master of none. Nat Rev Cancer 9: 821–829.
Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K et al. (2008). Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem 283: 9113–9126.
Lee H, Habas R, Abate-Shen C . (2004). MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304: 1675–1678.
Lennox RW, Cohen LH . (1984). The H1 Subtypes of Mammals: Metabolic Characteristics and Tissue Distribution. John Wiley & Sons: New York.
Lever MA, Th'ng JP, Sun X, Hendzel MJ . (2000). Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408: 873–876.
Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W . (2004). Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA 101: 2259–2264.
Ma Y, Lieber MR . (2006). In vitro nonhomologous DNA end joining system. Methods Enzymol 408: 502–510.
McBryant SJ, Lu X, Hansen JC . (2010). Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res 20: 519–528.
Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand S . et al. (2004). Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell 13: 251–263.
Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL . (2006). Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev 20: 1959–1973.
Parseghian MH, Hamkalo BA . (2001). A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem Cell Biol 79: 289–304.
Roth SY, Allis CD . (1992). Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci 17: 93–98.
Toledo F, Wahl GM . (2006). Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923.
Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D . (2004). Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16: 93–105.
Vila R, Ponte I, Jimenez MA, Rico M, Suau P . (2000). A helix-turn motif in the C-terminal domain of histone H1. Protein Sci 9: 627–636.
Vogelstein B, Lane D, Levine AJ . (2000). Surfing the p53 network. Nature 408: 307–310.
Woodcock CL, Skoultchi AI, Fan Y . (2006). Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14: 17–25.
Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S et al. (2010). Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol 189: 407–415.
Acknowledgements
We acknowledge Michael Lieber for DNA-PK and Daniela Rhodes for p601-7. This research was supported by NIH-R01GM84209 (WA), ACS RSG DMC-1005001 (WA) and NIH-RO1DK04393 (MRS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, K., Jeong, K., Kim, H. et al. Functional interplay between p53 acetylation and H1.2 phosphorylation in p53-regulated transcription. Oncogene 31, 4290–4301 (2012). https://doi.org/10.1038/onc.2011.605
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.605
Keywords
This article is cited by
-
An Arabidopsis Linker Histone-Like Protein Harbours a Domain with Adenylyl Cyclase Activity
Plant Molecular Biology Reporter (2023)
-
Emerging roles of linker histones in regulating chromatin structure and function
Nature Reviews Molecular Cell Biology (2018)
-
Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair
Cell Research (2018)
-
Upregulation of the ALDOA/DNA-PK/p53 pathway by dietary restriction suppresses tumor growth
Oncogene (2018)
-
Signaling coupled epigenomic regulation of gene expression
Oncogene (2017)