Abstract
The epidermal growth factor receptor (EGFR), a member of the ErbB family of receptor tyrosine kinases, is expressed in up to 70% of epithelial ovarian cancers (EOCs), where it correlates with poor prognosis. The majority of EOCs are diagnosed at an advanced stage, and at least 50% present malignant ascites. High levels of IL-6 have been found in the ascites of EOC patients and correlate with shorter survival. Herein, we investigated the signaling cascade led by EGFR activation in EOC and assessed whether EGFR activation could induce an EOC microenvironment characterized by pro-inflammatory molecules. In vitro analysis of EOC cell lines revealed that ligand-stimulated EGFR activated NFkB-dependent transcription and induced secretion of IL-6 and plasminogen activator inhibitor (PAI-1). IL-6/PAI-1 expression and secretion were strongly inhibited by the tyrosine kinase inhibitor AG1478 and EGFR silencing. A significant reduction of EGF-stimulated IL-6/PAI-1 secretion was also obtained with the NFkB inhibitor dehydroxymethylepoxyquinomicin. Of 23 primary EOC tumors from advanced-stage patients with malignant ascites at surgery, 12 co-expressed membrane EGFR, IL-6 and PAI-1 by immunohistochemistry; both IL-6 and PAI-1 were present in 83% of the corresponding ascites. Analysis of a publicly available gene-expression data set from 204 EOCs confirmed a significant correlation between IL-6 and PAI-1 expression, and patients with the highest IL-6 and PAI-1 co-expression showed a significantly shorter progression-free survival time (P=0.028). This suggests that EGFR/NFkB/IL-6-PAI-1 may have a significant impact on the therapy of a particular subset of EOC, and that IL-6/PAI-1 co-expression may be a novel prognostic marker.
Similar content being viewed by others
Introduction
Epithelial ovarian cancer (EOC) is one of the most lethal gynecological cancers. Metastatic dissemination of EOC is confined to the intraperitoneal cavity and involves malignant ascites (Bast et al., 2009). Its specific metastatic pattern of spread suggests the presence of specific microenvironmental factors that guide the formation of permissive niches that are very important for the growth and development of secondary lesions. EOCs, as other solid tumors, are strictly associated with inflammation and a complex cytokine/chemokine network. By computational search of co-expression of ligand/receptor pairs in EOC gene-expression data sets, our group has highlighted the possible signaling activated by specific chemokines (Castellano et al., 2006), whereas others have reported that the receptor/ligand pair CXCR4/CXCL12 and the cytokines IL-1β, IL-6 and IL-8 contribute to proliferation of EOC (Hagemann et al., 2006). EOC cells in vitro have also been demonstrated to modulate the macrophage phenotype by inducing the expression of inflammatory mediators (Hagemann et al., 2005). Among the cyto/chemokines found in EOC ascites, IL-6 has been shown to be a growth-promoting and anti-apoptotic factor (Kryczek et al., 2000). Moreover, patients with high levels of IL-6 expression have shorter survival than patients with lower levels (Penson et al., 2000). Metastatic and drug resistant recurrent EOC have significantly higher IL-6 production compared with that in primary tumors. Indeed, treatment of EOC cell lines with a chimeric anti-IL-6 monoclonal antibody increased paclitaxel cytotoxicity (Guo et al., 2010).
The connection between inflammation and cancer consists of extrinsic and intrinsic pathways, driven by inflammatory conditions and genetic alterations (for example, mutations in genes encoding RAS, MYC and RET), respectively. The epidermal growth factor receptor (EGFR), a member of ErbB family of receptor tyrosine kinases, activates multiple signaling cascades that cause growth and invasion of tumor cells. In most cancers, its expression has been associated with disease progression and poor outcome (Lemmon and Schlessinger, 2010). Although EGF-stimulated EOC cells that undergo epithelial–mesenchymal transition may upregulate production of IL-6 (Colomiere et al., 2009), the EGFR signaling cascade has not yet been directly associated with induction of an inflammatory network. In a homeostatic state, EGFR has a key role in normal ovarian follicle development and cell-growth regulation of the ovarian surface epithelium (Conti et al., 2006), whose cells might give rise to EOC. In EOC tumor samples, EGFR is expressed in an estimated 10–70% of EOCs, and its altered expression is associated with advanced-stage disease and poor prognosis (Hudson et al., 2009). Very recently, EGFR overexpression has been also associated with a lack of response to chemotherapy in patients with EOC (Sheng and Liu, 2011). EGFR is considered to be a key therapeutic target in many types of cancer. However, for reasons that are still unclear, treatment of EOC patients with anti-EGFR results in a very poor response and is not correlated to EGFR expression (Siwak et al., 2010). Therefore, there is an urgent need to further understand the relationships between the tumor microenvironment, EGFR activation and disease outcome in ovarian cancer.
Herein, we further investigate the signaling cascade led by EGFR activation in EOC, and assess whether EGFR activation may induce a microenvironment that favors survival of EOC by stimulating the expression of IL-6 and other pro-inflammatory molecules. In vitro, it was found that ligand-dependent EGFR activation triggered co-expression of the pro-inflammatory molecules IL-6 and plasminogen activator inhibitor-1 (PAI-1) via transcriptional activation of NFkB. High levels of IL-6 and PAI-1 co-expression were also found to characterize a subset of advanced EOC that express membrane EGFR.
Results
Ligand-dependent EGFR activation is not a predictor of sensitivity to anti-EGFR compounds
EOC cell lines were analyzed for EGFR expression by flow cytometry; all those derived from patients with serous adenocarcinomas expressed variable levels of membrane EGFR (Supplementary Figure 1A). In the same cell lines, IL-6 release was determined, in media conditioned for 24 h, by ELISA. IGROV1 and OAW42 cell lines released the highest amounts of IL-6 (422 pg/ml and 140 pg/ml, respectively) (Supplementary Figure 1B) and were selected for further characterization.
IGROV1 and OAW42 cells stimulated with EGF up to 60 min were analyzed by western blotting for the EGFR signaling cascade. In both the cell lines, phospho-EGFR was detected after 5 min of EGF stimulation reaching a maximum after 60 min. However, EGFR appeared to be degraded within a few minutes in IGROV1 cells (Supplementary Figure 1C), whereas it remained stable in OAW42, suggesting that the receptor is immediately degraded upon stimulation only in the former. EGFR phosphorylation was assessed in cells stimulated for 20 min with EGF alone or together with the tyrosine kinase inhibitor AG1478. In both cell lines, AG1478 completely inhibited EGFR phosphorylation together with that of the downstream effector ERK and most AKT phosphorylation (Figure 1a).
Ligand-dependent EGFR activation is not a predictor of sensitivity to anti-EGFR compounds. (a) Western-blot analysis was performed on total lysates from starved IGROV1 and OAW42 cells left untreated or treated for 20 min with EGF alone or together with AG1478. The antibodies used are indicated. β-actin is shown as a control for protein loading. A representative experiment is shown; all experiments were performed in triplicate. (b) IGROV1 and OAW42 cells were exposed to different concentrations of cetuximab for 96 h in medium with fetal calf serum. (c) Dose-response curves of IGROV1 and OAW42 cells treated with Erlotinib or Gefitinib for 72 h. (b, c): Representative growth curves of three independent experiments are shown; each point represents the mean of five independent replicates±s.d. Asterisks indicate a significant difference by two-way analysis of variance.
The effects on cell growth of the anti-EGFR monoclonal antibody cetuximab, which affects the ligand binding to the receptor, and the EGFR inhibitors gefitinib and erlotinib, which interfere with ATP for binding to EGFR, were then evaluated. Although EGFR in both cell lines is activated by the ligand, only the proliferation of IGROV1 cells (50% inhibition with 5 μg/ml), but not that of OAW42 cells (15% inhibition with 100 μg/ml), was affected by cetuximab treatment (Figure 1b). Accordingly, IGROV1 cells were about eight-fold more sensitive to gefitinib and erlotinib than OAW42 cells (Figure 1c).
Ligand-dependent EGFR signaling triggered IL-6 production and NFkB activation
IL-6 release, quantified in conditioned media after EGF stimulation alone or together with AG1478, was time dependent for both cell lines; after 24 h it was almost completely inhibited by AG1478 treatment in IGROV1 cells, but only partially inhibited in OAW42 cells (96% and 30%, respectively) (Figure 2a). In starved IGROV1 cells, EGFR appeared to be directly involved in IL-6 release, as its release was inhibited by AG1478, suggesting autocrine EGFR activation. In starved OAW42 cells, IL-6 production was not inhibited by AG1478 treatment, and EGF stimulation led to IL-6 levels that were seven-fold higher compared with fetal calf serum-maintained cells (see Supplementary Figure 1B), arguing for a mechanism of IL-6 production that is independent of EGFR activation.
Ligand-dependent/EGFR signaling triggers IL-6 production via NFkB activation. (a) IL-6 levels measured by ELISA on conditioned media from starved EOC cell lines left untreated, or treated for the indicated times with EGF (20 ng/ml) alone or together with AG1478 (20 μM). Representative growth curves of one of three independent experiments are shown. Each point represents the mean of five independent replicates ± s.d. (b) Real-time PCR for IL-6 from total RNA of starved EOC cell lines left untreated, treated for 24 h with EGF (20 ng/ml) alone or together with AG1478 (20 μM), LY294002 (50 μM) or with UO126 (40 μM). Results are presented as relative expression normalized to GAPDH mRNA levels. (c) Luciferase promoter gene assay of starved EOC cells transiently transfected with reporter plasmids containing the NFkB binding sites and stimulated for 24 h with EGF (20 ng/ml). (d) Inhibition of NFkB transcriptional activity of starved EOC cells transiently transfected as above and stimulated for 24 h with EGF alone or together with AG1478 (black), LY294002 (gray) and UO126 (white). (c, d) Data are mean values (±s.d.) normalized for transfection efficiency in a representative experiment.
In agreement with this possibility, IL-6 transcript levels evaluated by real-time RT–PCR increased about 20-fold after 24 h of stimulation with EGF in IGROV1 cells, and were inhibited to levels comparable to those in starved cells after treatment with AG1478, as well as with the PI3K inhibitor LY294002 and the MEK inhibitor UO126; in OAW42 cells, IL-6 transcript levels increased only about three-fold and were not inhibited by AG1478. LY294002 treatment inhibited the expression of IL-6 transcripts, whereas UO126 led to a 16-fold increase in IL-6 transcript levels (Figure 2b).
NFkB activity, which is known to activate the transcription of inflammation-related proteins such as IL-6 (Karin, 2006), was then assayed upon EGF stimulation by transient transfection with a luciferase reporter construct containing two NFkB-binding sites. In IGROV1 cells, NFkB-associated promoter activity increased within 1 h, reaching a maximum after 3 h EGF stimulation and remained stable up to 24 h. In contrast, in OAW42 cells NFkB-containing promoter activity was measurable only after 20 h of EGF stimulation (Figure 2c). In order to evaluate whether NFkB-containing promoter activity was dependent on the EGFR signaling cascade, it was measured in the presence of AG1478, LY294002 or UO126. EGF-stimulated NFkB-containing promoter activity was almost completely inhibited by AG1478 and UO126 treatment, strongly reduced (60%) by LY294002 treatment in IGROV1 cells (Figure 2d), and inhibited about 40%, 80% and 65% with AG1478, LY294002 and UO126 treatment, respectively, in OAW42 cells.
These results demonstrate that, in IGROV1 cells, but not in OAW42, ligand-dependent EGFR/MEK/ERK or EGFR/PI3K/AKT activation leads to transcriptional activation of NFkB and IL-6 production.
Ligand-dependent EGFR activation and expression induced production of specific cyto/chemokines
To evaluate whether EGF stimulation of EOC cells could induce the expression and release of other inflammatory cyto/chemokines in addition to IL-6, we performed a 51 cyto/chemokine multiplex analysis using Bioplex technology on IGROV1 conditioned media collected after EGF stimulation alone or together with AG1478. In media collected after 24 h of EGF stimulation, several soluble factors were found to be upregulated and inhibited by concomitant treatment with AG1478, namely IL-6, vascular endothelial growth factor, PAI-1, TGF (transforming growth factor) α, macrophage colony stimulating factor and interleukin-8 (IL-8) (Supplementary Table 2). Figure 3a shows the time course of levels of the soluble factors, PAI-1, IL-8 and IL-6 (Figure 2a), which all exhibited a two-fold increase after 24 h of EGF stimulation, and whose release was inhibited by AG1478 treatment.
Ligand-dependent EGFR activation induced the production of specific cyto-/chemokines. (a) Fold change in PAI-1 and IL-8 levels compared with unstimulated cells released in media from starved IGROV1 cells stimulated for 4, 8 and 24 h with EGF alone (filled square), EGF plus AG1478 (open square), or AG1478 alone (open circle) evaluated by the Procarta cytokine assay. (b) Real-time RT–PCR for PAI-1 and IL-8 from total RNA of IGROV1 treated as above. Results are presented as relative expression normalized for GAPDH mRNA levels. (c) IL-6, PAI-1 and IL-8 levels measured by the Procarta cytokine assay on conditioned media from starved EOC cell lines left untreated, treated for 24 h with EGF alone, or together with DHMEQ (5 μg/ml). Asterisks indicate a significant difference by one-way analysis of variance (b) and t-test (c).
As observed for IL-6 transcripts (Figure 2b), PAI-1 and IL-8 transcript levels were upregulated after 24 h of EGF stimulation and inhibited by treatment with AG1478 (Figure 3b).
To evaluate the involvement of NFkB transcriptional activity in the production of IL-6, PAI-1 and IL-8, the Bioplex quantification was also applied to conditioned media after EGF stimulation alone or together with Dehydroxymethylepoxyquinomicin (DHMEQ). DHMEQ decreased the levels of the IL-6 (from 250 to 217 pg/ml), PAI-1 (from 366.51 to 254.8 pg/ml) and IL-8 (from 26.6 to 15.5 pg/ml) secreted by starved cells, suggesting an autocrine activity. A further and significant decrease of IL-6 (from 500.4 to 341.8 pg/ml, 32%), PAI-1 (from 1225.76 to 807.29 pg/ml, 35%) and IL-8 (from 54.94 to 30.29 pg/ml, 45%) releases was observed upon DHMEQ treatment of EGF-stimulated cells. These data argued for the notion that in EGF-stimulated IGROV1 cells IL-6, PAI-1 and IL-8 productions are partially dependent on NFkB transcriptional activation.
IL-6 and PAI-1 expressions are inhibited by EGFR knockdown
To validate these results, EGFR knockdown was performed with anti-EGFR small-interfering RNA (siRNA). Forty-eight hours after transfection, total cell lysates from silenced IGROV1 cells were analyzed by western blotting to evaluate EGFR expression and phosphorylation of the downstream effectors AKT and ERK. A 60% reduction in EGFR expression corresponded to 90% and 50% reduction of ERK and AKT phosphorylation, respectively (Figure 4a). NFkB transcriptional activity of EGFR-silenced cells was 70% lower compared with cells transfected with a control siRNA (Figure 4b). Real-time RT–PCR on total RNA from EGFR-silenced cells revealed that a 15-fold decrease of EGFR transcript was accompanied by a dramatic decrease of PAI-1 and IL-6 transcripts, and to only a 25% decrease in IL-8 transcripts (Figure 4c). The decrease in IL-6 transcript levels was similar to that obtained in IGROV1 cells stimulated with EGF and treated with AG1478 (Figure 2b), demonstrating a direct correlation between EGFR expression/activation and IL-6/PAI-1 transcription and production.
IL-6 and PAI-1 expressions are inhibited by EGFR knockdown. (a) Western-blot analysis of total lysates from IGROV1 cells treated with control or EGFR-specific siRNA. The antibodies used are indicated. β-actin is shown as a control for protein loading. A representative experiment is shown. (b) Luciferase promoter gene assay of EGFR-silenced IGROV1 cells transiently transfected with reporter plasmids containing NFkB binding sites. Data are mean values (±s.d.) normalized for transfection efficiency in three independent experiments performed in triplicate. (c) Real-time RT–PCR analysis for EGFR, PAI-1, IL-6 and IL-8 on total RNA from EGFR-silenced IGROV1. Results are represented as relative mRNA expression normalized for GAPDH mRNA levels. (b, c): Asterisks indicate a significant difference by t-test.
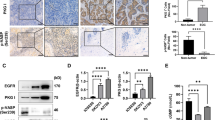
IL-6 and PAI-1 are highly co-expressed in EOC samples
To validate the in vitro results, we used immunohistochemistry to evaluate the expression of EGFR, IL-6 and PAI-1 in 23 formalin-fixed, paraffin-embedded primary ovarian cancers (Table 1). The characteristics of patients and cytological information associated with the ascites are summarized in Supplementary Table 1. The majority of EOC samples (74%) heterogeneously expressed EGFR on the membrane of tumor cells; samples with the strongest reactivity with the anti-EGFR antibody also showed EGFR expression in the cytoplasm. Six samples stained with anti-EGFR only in the cytoplasm. Tumor cells of 16 of 23 EOC samples (70%) also showed staining with the anti-IL-6 antibody. Of the 23 samples with the highest EGFR expression, 4 also showed the strongest staining with anti-IL-6 (for example, sample no. 22, Figure 5a). In some sections with high reactivity for anti-EGFR, not all tumor cells stained with anti-IL-6. Some samples showed homogenous staining with anti PAI-1 (samples no. 7 and no. 23), whereas in other samples PAI-1 expression appeared heterogeneous. Interestingly, the samples that stained strongly with anti PAI-1 were also strongly reactive with both anti-EGFR and -IL-6 antibodies (Figure 5a).
IL-6 and PAI-1 are highly co-expressed in EOC samples. (a) Immunohistochemistry for EGFR, IL-6 and PAI-1 in a primary formalin-fixed, paraffin-embedded EOC sample (characteristics reported in Supplementary Table 1). Representative images from sample no. 22 are shown. (b) IL-6, PAI-1 and IL-8 quantification by ELISA in 23 ascites samples from the same EOC patients. (c) Correlation between IL-6 and PAI-1 levels measured in ascites. The linear regression line, Spearman correlations and P-values are shown.
IL-6 and PAI-1 levels were measured in the corresponding ascites. Medium/high levels of both IL-6 and PAI-1 were seen in 12 of 23 ascites (Figure 5b), and their expression showed a positive correlation (r=0.68; P=0.002) (Figure 5c). These 12 EOC ascites samples were derived from patients whose primary tumors co-expressed membrane EGFR, IL-6 and PAI-1. In contrast, the presence of IL-8 in ascites was not associated with release of IL-6 or PAI-1, although some samples contained detectable levels of the three molecules. Furthermore, in ascites the majority of samples with medium/high IL-6 and PAI-1 expression contained tumor cells alone or together with immune cells (Supplementary Table 1 and Supplementary Figure 3), excluding that the two molecules could be released only by immune cells.
Expression of EGFR-associated molecules in publicly available EOC data sets
To investigate the relationship between EGFR, IL-6, PAI-1 and IL-8 expression, we analyzed four publicly available data sets containing the gene-expression profile of EOC patients. From data set I (Supplementary Table 3), the profiles of 204 serous EOC were selected and the expression intensity of the EGFR, IL-6, PAI-1 and IL-8 genes were plotted together to evaluate trends in expression. EGFR transcript levels appeared to be highly homogeneous, and no significant correlations were found (Supplementary Figure 2A). The highest correlation score (r=0.58, P<0.0001) was found between IL-6 and PAI-1; all the other combinations showed lower correlation scores ranging from r=0.44 (for IL-8 and IL-6) to r=0.30 (for IL-8 and PAI-1) (Figure 6a). A significant correlation between IL-6 and PAI-1 was also obtained after analysis of the other three data sets (Supplementary Figure 2B).
Expression of EGFR-associated molecules in EOC data set I. (a) Correlation among IL-6, PAI-1 and IL-8 were determined and the values are plotted on a log2 scale. Pearson correlations (r), linear regression and P‐values are reported. (b) Association between IL-6/PAI-1 expression intensities and tumor stage was analyzed on samples filtered for expression higher or lower than the respective median. Asterisk indicates a significant difference by Fisher's exact test. (c) Kaplan–Meier curves reporting the progression-free survival analysis on patient subgroups selected for IL-6/PAI-1 expression higher than the 3rd quartiles and lower than the 1st quartile (log-rank test).
Upon filtration of data set I for high or low expression of both IL-6 and PAI-1, it was found that high co-expression of IL-6 and PAI-1 was significantly associated with the advanced-stage EOC (Fisher's exact test, P=0.0006) (Figure 6b). In the same data set, progression-free survival was analyzed in the subset of patients exhibiting expression levels of both IL-6 and PAI-1 below the first and above the third quartile. High IL-6 and PAI-1 was significantly associated with shorter progression-free survival (log-rank test, P=0.028; HR=2.13, confidence interval=1.08–4.20) (Figure 6c). Of note, the expression of either IL-6 or PAI-1 when considered individually did not discriminate between the two groups.
Discussion
This study showed that IL-6 and PAI-1 co-expression is a potential marker for the ligand-dependent EGFR/NFkB signaling cascade. In vitro we demonstrated that the ligand-dependent EGFR/NFkB signaling cascade leads to co-expression of IL-6 and PAI-1. Ex vivo, using 23 EOC from advanced-stage patients with malignant ascites at surgery, we observed co-expression of EGFR, IL-6 and PAI-1 in 57% of primary tumors, and concomitant expression of both IL-6 and PAI-1 in the corresponding ascites. Finally, in-silico analysis on four publicly available data sets of EOC gene expression showed a correlation between the expression of the IL-6 and PAI-1 genes in advanced EOC patients, which in one case was associated with shorter progression-free survival.
Despite the evidence for EGFR expression in the majority of EOCs, to date a uniform picture of the biological and clinical consequences of EGFR expression and activation has not emerged. In advanced EOCs, the expression of EGFR and PAI-1(Carey et al., 2010), or IL-6 (Guo et al., 2010) are associated with chemoresistance, thus supporting the hypothesis of a functional connection between these three molecules in the progression and chemoresistance of advanced EOCs.
In the present study, we provide several lines of evidence suggesting that simply considering EGFR expression levels alone is not sufficient to define a role of receptor activation. We propose here that co-expression as well as the concomitant presence of IL-6 and PAI-1 in EOC ascites could characterize a subset of EGFR-expressing EOCs with shorter progression-free survival after chemotherapy. As we have also shown in vitro that the EOC cell line resembling this subset of EOC is more sensitive to anti-EGFR compounds, it can be argued that the association of anti-EGFR agents with taxol and cisplatin could be more appropriate for treating this subset of EOCs.
In EOC cells, the mitogenic effects of EGFR activation has been already documented and leads to upregulation of genes involved in cell cycle and proliferation, apoptosis and protein turnover (Siwak et al., 2010). The present in vitro data demonstrate that a signaling cascade from ligand-stimulated EGFR/MEK/ERK or EGFR/PI3K/AKT activates the transcriptional activity of NFkB with expression of IL-6 together with PAI-1. A similar signaling pathway has been experimentally demonstrated in glioblastoma cell lines (Paugh et al., 2008), and a positive correlation between PAI-1 and IL-6 gene expression has been observed in a meta-analysis carried out on publicly available data sets of gene expression of breast carcinoma (Sternlicht et al., 2006). Interestingly, very recently ErbB2 expression, another member of the ErbB family, was found to upregulate IL-6 production through the transcriptional activity of NFkB (Hartman et al., 2011).
In vitro, we found that the EGFR/NFkB signaling cascade is ligand dependent in an in vitro EOC model, which is in agreement with previous reports on EGFR activation by ligands, such as EGF, TGFα and the heparin-binding-EGF (Hudson et al., 2009) present in the EOC microenvironment. EGFR ligands can be found in the EOC microenvironment due to the activation of A disintegrin and metalloprotease by endothelin-1- or lysophosphatidic acid-stimulated G-protein-coupled receptors (Braun and Coffey, 2005). Several growth factors including EGF may also induce activation of NFkB. NFkB is a family of transcription factors that modulate immunological and inflammatory responses by directly controlling the transcription of pro-inflammatory cytokines. NFkB is activated in many malignancies including ovarian cancer (Karin, 2006). Production of IL-6 has been shown to be increased in the EOC cell lines SKOV3 and OVCA433 after stimulation with EGF, as observed here, although the molecular mechanism responsible for the increase is still unclear (Colomiere et al., 2009). In ER-negative breast carcinoma, stimulation with EGF activates NFkB through an unknown mechanism that may involve the scaffold protein Carma3 (Van Laere et al., 2007). Indeed, Carma3 has been recently demonstrated to be a link between EGFR, Ikk kinase and NFkB activation (Jiang et al., 2011). Further experiments are needed to confirm or exclude that this mechanism is linked with EGFR activation of NFkB transcriptional activity.
Our demonstration that the levels of IL-6 and PAI-1 transcripts and proteins correlate with one another may be advantageous not only in studying the mechanisms associated with progression of EOC, but also for their exploitation as potential prognostic markers. PAI-1, one of three plasminogen-activator inhibitors, is a major regulator of the pericellular plasmin-generating uPA/uPAR cascade. To date, both uPA and uPAR are associated with invasiveness and metastasis of a variety of cancers, including EOC (Dass et al., 2008). In breast carcinoma, uPA/PAI-1 expression levels have provided clinically relevant information on the risk of relapse and may have relevance when choosing endocrine therapies and chemotherapy (Leissner et al., 2006); PAI-1 has been associated with shorter recurrence-free and overall survival, although its expression is rarely used as prognostic/predictive factor, as protein-based assays are difficult to routinely adopt to the limited amount of biopsy tissue that is generally available (Look et al., 2002). In contrast, in EOC PAI-1 expression in ascites could be more easily tested compared with other tumors. Furthermore, in silico analysis, performed on a very informative data set of EOC gene expression that reports detailed clinical annotations and used by other investigators to validate high-throughput results (Cancer Genome Atlas Research Network, 2011), gives a clear indication that also the use of concomitant IL-6 and PAI-1 gene expression may be a useful tool to identify EOC patients whose tumors are potentially resistant to conventional chemotherapy. A study on a large number of EOC patients is therefore warranted to further validate these data.
Although further study of EGFR-mediated inflammation in EOC is needed, taken together these findings highlight novel and interesting tools that may provide useful therapeutic information for a particular subset of EGFR-expressing EOC and the use of IL-6 and PAI-1 co-expression as a potential prognostic marker.
Materials and methods
Tumor samples
The Institutional Review Board approved the use of archived material and ascites, as well as clinical data. All clinical specimens were accompanied by informed consent from all patients to use excess biological material for investigative purposes. Histological selection of patients was based on advanced stage at diagnosis and the presence of ascites at surgery. EOC samples were selected and collected by a pathologist (BV). Twenty-three pairs of formalin-fixed, paraffin-embedded primary tumors and ascites were used.
Antibodies and reagents
The following antibodies were used in blotting experiments: anti-EGFR, anti-MAP kinase activated (pERK1/2), anti-AKT, anti-phosphoAKT (Ser473) (clone D9E) from Cell Signaling Technology (New England BioLabs, Beverly, MA, USA); anti-phospho-EGFR (Tyr1173) from Nano Tools (Teningen, Germany); anti-MAP kinase (ERK1/2) from Santa Cruz Biotecnology (Santa Cruz, CA, USA) and anti-Actin from Sigma-Aldrich (St Louis, MO, USA). The anti-EGFR monoclonal antibody MINT-5 was used for flow cytometry (Tosi et al., 1995). Fluorochrome-conjugated Alexa Fluor 488 secondary Ab was from Molecular Probes (Invitrogen, San Francisco, CA, USA). AG1478 was from Sigma-Aldrich. Gefitinib and erlotinib were from Axon MedChem (Groningen, the Netherlands); the MEK inhibitor UO126 was from Promega (Promega, Madison, WI, USA) and the PI3K inhibitor LY294002 was from Sigma Aldrich. DHMEQ, an NFkB inhibitor that blocks the binding of NFkB to DNA (Umezawa, 2006; Yamamoto et al., 2008), was synthesized by one of the authors (KU). Recombinant human EGF was from Peprotech (Peprotech Inc, Rocky Hill, NJ, USA). Erbitux (Merck-Serono, Geneva, Switzerland) was used for cetuximab. Taqman Gene Expression Assays were from Applied Biosystems (Foster City, CA, USA). ELISA for IL-6, PAI-1 and IL-8 dosage was from R&D Systems (Minneapolis, MN, USA).
Immunohistochemistry
Detailed immunohistochemistry studies are reported in the Supplementary Materials and Methods.
Ovarian cancer cell culture
OVCAR3, OVCA432, SKOV3, IGROV1 (serous histotype) and A2780 (mucinous histotype) cell lines were maintained in RPMI 1640 (Sigma Aldrich) with 10% fetal calf serum (Hyclone, Logan, UT, USA) and 2 mmol/l glutamine in a 5% CO2 humidified atmosphere at 37 °C. OAW42 (serous histotype, kindly provided by Dr A. Ullrich, Max Planck Institute of Biochemistry, Martinsried, Germany) cells were cultured in MEM (Sigma Aldrich) and supplemented as above.
ELISA
Cell lines were grown until confluence, fresh medium was added and samples for IL-6 dosage were collected after 24 h.
Western blotting
Preparation of total cell lysates and Western blotting analysis was performed as previously described (De Santis et al., 2009). Blots were viewed and analyzed using ChemiDoc XRS and Quantity One software (Biorad, Hercules, CA, USA).
Cell proliferation
IGROV1 and OAW42 cells were plated into 96-well plates at a density of 5 × 103 cells/well or 2.5 × 103 cells/well, respectively. At 0, 24, 48, 72 and 96 h, mitochondrial activity was measured using a CellTiter-Glo luminescent cell viability assay performed according to the manufacturer's instructions (Promega).
Analysis of promoter activity
Analysis of promoter activity was performed as described (Tomassetti et al., 2008). Cells were transfected with a plasmid containing the NFkB promoter–reporter gene construct (Promega) using Lipofectamine 2000 (Invitrogen). At 24 h after transfection, cells were inhibited with AG1478, LY294002, UO126 and/or stimulated with EGF 20 ng/ml (Peprotech). The dual-luciferase assay was performed as suggested by the manufacturer (Promega).
Cyto/chemokines quantification in cell culture media
Conditioned media from IGROV1 cells was collected after 4, 8 and 24 h of EGF stimulation alone or together with AG1478 (20 μM) or with the NFkB inhibitor DHMEQ (5 μg/ml). The samples and an aliquot of the medium were analyzed in duplicate. The assay was performed using the Procarta cytokine kits on the Luminex platform (Affimetrix, Santa Clara, CA, USA) as described in the Supplementary Materials and Methods.
RNA extraction and real-time RT–PCR analysis
Total RNA from cell lines was extracted using a commercial kit (Amersham Bioscience-GE Healthcare, Piscataway, NJ, USA). RT–PCR analysis was performed as described (Degl’Innocenti et al., 2010).
siRNA treatment
Cells were transfected with 40 pmol/ml siRNA duplex against EGFR mRNA (Smart Pool, Thermo Scientific, Dharmacon Inc, Chicago, IL, USA) or control siRNA (Quiagen-Xeragon, Germantown, MD, USA). Transfection was performed using Lipofectamine 2000, according to the manufacturer's protocol. Total RNA and lysates were prepared at 48 h after transfection.
Bioinformatic and statistical analyses
Four publicly available data sets of gene expression generated using the Affymetrix platform were downloaded from the web (see Supplementary Table 3) (Bild et al., 2006; Anglesio et al., 2008; Tothill et al., 2008; Berchuck et al., 2009) and their characteristics are reported in the Supplementary Materials and Methods. Before analysis, samples of data set I (GSE9899) were filtered for ‘malignant’, primary site ‘ovary’ and histological type ‘serous’, and the remaining 204 samples were used. Data were normalized by the RMA algorithm. Cross-hybridizing probes were filtered out and data were collapsed on Gene Symbol using the median of probes. EGFR, IL-6, SERPINE1 (the gene encoding for PAI-1, as named herein) and IL-8 gene-expression intensities were extracted and analyzed further. The Pearson correlation scores with the related P-value (two-tailed) were computed for each gene pair.
Patients in data set I were categorized as expressing both IL-6 and PAI-1 higher (n=77) or lower (n=78) than the respective median and divided in ‘early’ (I and II, n=15) and ‘advanced’ (III and IV, n=140) stages.
The survival analysis was performed in data set I by considering only patients with both IL-6 and PAI-1 expression intensities below the first (n=22) and above the third (n=28) quartile. Progression-free survival (defined as the time interval between the date of diagnosis and the first confirmed sign of disease recurrence) was used as the primary end point. Curves were generated with the Kaplan Meyer method, and hazard ratios and 95% confidence intervals were also computed.
GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA), R statistical language version 2.10.0 (URL http://www.R-project.org) and Bioconductor (URL http://www.bioconductor.org) were used for statistical tests. The P-values of all statistical tests were two-sided; a P⩽0.05 was considered significant. Significance of differences was determined: for in vitro assays, by one- and two-way analysis of variance and Student's t-test when appropriate; for association between categorical variables by Fisher's exact test; for survival analysis with a non-parametric (log-rank) test.
References
Anglesio MS, Arnold JM, George J, Tinker AV, Tothill R, Waddell N et al. (2008). Mutation of ERBB2 provides a novel alternative mechanism for the ubiquitous activation of RAS-MAPK in ovarian serous low malignant potential tumors. Mol Cancer Res 6: 1678–1690.
Bast Jr RC, Hennessy B, Mills GB. (2009). The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 9: 415–428.
Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA et al. (2009). Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin Cancer Res 15: 2448–2455.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439: 353–357.
Braun AH, Coffey RJ. (2005). Lysophosphatidic acid, a disintegrin and metalloprotease-17 and heparin-binding epidermal growth factor-like growth factor in ovarian cancer: the first word, not the last. Clin Cancer Res 11: 4639–4643.
Carey MS, Agarwal R, Gilks B, Swenerton K, Kalloger S, Santos J et al. (2010). Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin Cancer Res 16: 2852–2860.
Castellano G, Reid JF, Alberti P, Carcangiu ML, Tomassetti A, Canevari S. (2006). New Potential Ligand-Receptor Signaling Loops in Ovarian Cancer Identified in Multiple Gene Expression Studies. Cancer Res 66: 10709–10719.
Colomiere M, Ward AC, Riley C, Trenerry MK, Cameron-Smith D, Findlay J et al. (2009). Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer 100: 134–144.
Conti M, Hsieh M, Park JY, Su YQ. (2006). Role of the epidermal growth factor network in ovarian follicles. Mol Endocrinol 20: 715–723.
Dass K, Ahmad A, Azmi AS, Sarkar SH, Sarkar FH. (2008). Evolving role of uPA/uPAR system in human cancers. Cancer Treat Rev 34: 122–136.
De Santis G, Miotti S, Mazzi M, Canevari S, Tomassetti A. (2009). E-cadherin directly contributes to PI3K/AKT activation by engaging the PI3K-p85 regulatory subunit to adherens junctions of ovarian carcinoma cells. Oncogene 28: 1206–1217.
Degl'Innocenti D, Alberti C, Castellano G, Greco A, Miranda C, Pierotti MA et al. (2010). Integrated ligand-receptor bioinformatic and in vitro functional analysis identifies active TGFA/EGFR signaling loop in papillary thyroid carcinomas. PLoS One 5: e12701.
Genome Atlas Research Network (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615.
Guo Y, Nemeth J, O′Brien C, Susa M, Liu X, Zhang Z et al. (2010). Effects of siltuximab on the IL-6-induced signaling pathway in ovarian cancer. Clin Cancer Res 16: 5759–5769.
Hagemann T, Wilson J, Burke F, Kulbe H, Li NF, Pluddemann A et al. (2006). Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J Immunol 176: 5023–5032.
Hagemann T, Wilson J, Kulbe H, Li NF, Leinster DA, Charles K et al. (2005). Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J Immunol 175: 1197–1205.
Hartman ZC, Yang XY, Glass O, Lei G, Osada T, Dave SS et al. (2011). HER2 overexpression elicits a proinflammatory IL-6 autocrine signaling loop that is critical for tumorigenesis. Cancer Res 71: 4380–4391.
Hudson LG, Zeineldin R, Silberberg M, Stack MS. (2009). Activated epidermal growth factor receptor in ovarian cancer. Cancer Treat Res 149: 203–226.
Jiang T, Grabiner B, Zhu Y, Jiang C, Li H, You Y et al. (2011). CARMA3 is crucial for EGFR-Induced activation of NF-kappaB and tumor progression. Cancer Res 71: 2183–2192.
Karin M. (2006). Nuclear factor-kappaB in cancer development and progression. Nature 441: 431–436.
Kryczek I, Grybos M, Karabon L, Klimczak A, Lange A. (2000). IL-6 production in ovarian carcinoma is associated with histiotype and biological characteristics of the tumour and influences local immunity. Br J Cancer 82: 621–628.
Leissner P, Verjat T, Bachelot T, Paye M, Krause A, Puisieux A et al. (2006). Prognostic significance of urokinase plasminogen activator and plasminogen activator inhibitor-1 mRNA expression in lymph node- and hormone receptor-positive breast cancer. BMC Cancer 6: 216.
Lemmon MA, Schlessinger J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134.
Look MP, van Putten WL, Duffy MJ, Harbeck N, Christensen IJ, Thomssen C et al. (2002). Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J Natl Cancer Inst USA 94: 116–128.
Paugh BS, Paugh SW, Bryan L, Kapitonov D, Wilczynska KM, Gopalan SM et al. (2008). EGF regulates plasminogen activator inhibitor-1 (PAI-1) by a pathway involving c-Src, PKCdelta, and sphingosine kinase 1 in glioblastoma cells. FASEB J 22: 455–465.
Penson RT, Kronish K, Duan Z, Feller AJ, Stark P, Cook SE et al. (2000). Cytokines IL-1beta, IL-2, IL-6, IL-8, MCP-1, GM-CSF and TNFalpha in patients with epithelial ovarian cancer and their relationship to treatment with paclitaxel. Int J Gynecol Cancer 10: 33–41.
Sheng Q, Liu J. (2011). The therapeutic potential of targeting the EGFR family in epithelial ovarian cancer. Br J Cancer 104: 1241–1245.
Siwak DR, Carey M, Hennessy BT, Nguyen CT, McGahren Murray MJ, Nolden L et al. (2010). Targeting the epidermal growth factor receptor in epithelial ovarian cancer: current knowledge and future challenges. J Oncol 2010: 568938.
Sternlicht MD, Dunning AM, Moore DH, Pharoah PD, Ginzinger DG, Chin K et al. (2006). Prognostic value of PAI1 in invasive breast cancer: evidence that tumor-specific factors are more important than genetic variation in regulating PAI1 expression. Cancer Epidemiol Biomarkers Prev 15: 2107–2114.
Tomassetti A, De Santis G, Castellano G, Miotti S, Mazzi M, Tomasoni D et al. (2008). Variant HNF1 Modulates Epithelial Plasticity of Normal and Transformed Ovary Cells. Neoplasia 10: 1481–1492.
Tosi E, Valota O, Negri DRM, Adobati E, Mazzoni A, Meazza R et al. (1995). Anti-tumor efficacy of an anti-epidermal growth factor receptor monoclonal antibody and its F(ab′)2 fragment against high- and low-EGFR-expressing carcinomas in nude mice. Int J Cancer 62: 643–650.
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S et al. (2008). Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 14: 5198–5208.
Umezawa K. (2006). Inhibition of tumor growth by NF-kappaB inhibitors. Cancer Sci 97: 990–995.
Van Laere SJ, Van der AI, Van den Eynden GG, van DP, Van Marck EA, Vermeulen PB et al. (2007). NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br J Cancer 97: 659–669.
Yamamoto M, Horie R, Takeiri M, Kozawa I, Umezawa K. (2008). Inactivation of NF-kappaB components by covalent binding of (-)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J Med Chem 51: 5780–5788.
Acknowledgements
We thank the gynecologic clinical staff and the biorepository personnel at the Fondazione IRCCS Istituto Nazionale dei Tumori, whose activity made this study possible; we thank Dr. Patrick Moore for English editing of the manuscript. Financial Support: Italian Association for Cancer Research (IG4608 to SC) and Italian Ministry of Health (Grant Alleanza contro il Cancro, ACC and Progetto Oncologico di Medicina Molecolare: i Tumori Femminili to SC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Alberti, C., Pinciroli, P., Valeri, B. et al. Ligand-dependent EGFR activation induces the co-expression of IL-6 and PAI-1 via the NFkB pathway in advanced-stage epithelial ovarian cancer. Oncogene 31, 4139–4149 (2012). https://doi.org/10.1038/onc.2011.572
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.572
Keywords
This article is cited by
-
Human papillomavirus 16 E6 promotes angiogenesis of lung cancer via SNHG1
Cell Biochemistry and Biophysics (2023)
-
Biological and clinical impact of membrane EGFR expression in a subgroup of OC patients from the phase IV ovarian cancer MITO-16A/MANGO-OV2A trial
Journal of Experimental & Clinical Cancer Research (2023)
-
Screening assessment of trimethoxy flavonoid and - (-)-epigallocatechin-3-gallate against formalin-induced arthritis in Swiss albino rats and binding properties on NF-κB-MMP9 proteins
Future Journal of Pharmaceutical Sciences (2021)
-
Combinatorial treatment with Gefitinib and Bay11-7085 sensitizes primary Gefitinib-resistant OSCC cells by influencing the EGFR- NFκB signaling axis
Medical Oncology (2021)
-
Current insights into the metastasis of epithelial ovarian cancer - hopes and hurdles
Cellular Oncology (2020)