Abstract
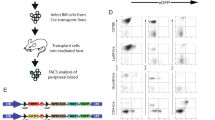
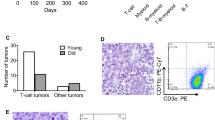
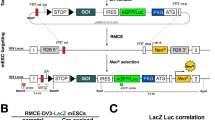
The Myc-deregulating chromosomal T(12;15)(Igh–Myc) translocation, the hallmark mutation of inflammation- and interleukin 6-dependent mouse plasmacytoma (PCT), is the premier model of cancer-associated chromosomal translocations because it is the only translocation in mice that occurs spontaneously (B lymphocyte lineage) and with predictably high incidence (∼85% of PCT), and has a direct counterpart in humans: Burkitt lymphoma t(8;14)(q24;q32) translocation. Here, we report on the development of a genetic system for the detection of T(12;15)(Igh–Myc) translocations in plasma cells of a mouse strain in which an enhanced green fluorescent protein (GFP)-encoding reporter gene has been targeted to Myc. Four of the PCTs that developed in the newly generated translocation reporter mice, designated iGFP5′Myc, expressed GFP consequent to naturally occurring T(12;15) translocation. GFP expression did not interfere with tumor development or the deregulation of Myc on derivative 12 of translocation, der (12), because the reporter gene was allocated to the reciprocal product of translocation, der (15). Although the described reporter gene approach requires refinement before T(12;15) translocations can be quantitatively detected in vivo, including in B lymphocyte lineage cells that have not yet completed malignant transformation, our findings provide proof of principle that reporter gene tagging of oncogenes in gene-targeted mice can be used to elucidate unresolved questions on the occurrence, distribution and trafficking of cells that have acquired cancer-causing chromosomal translocations of great relevance for humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Anderson PN, Potter M . (1969). Induction of plasma cell tumours in BALB-c mice with 2,6,10,14-tetramethylpentadecane (pristane). Nature 222: 994–995.
Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J et al. (2009). Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci USA 106: 2717–2722.
Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM . (1982). Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA 79: 7824–7827.
Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE et al. (2000). DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404: 510–514.
Eisenman RN . (2001). Deconstructing myc. Genes Dev 15: 2023–2030.
Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA et al. (2006). H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell 21: 201–214.
Germain RN, Castellino F, Chieppa M, Egen JG, Huang AY, Koo LY et al. (2005). An extended vision for dynamic high-resolution intravital immune imaging. Semin Immunol 17: 431–441.
Gostissa M, Ranganath S, Bianco JM, Alt FW . (2009a). Chromosomal location targets different MYC family gene members for oncogenic translocations. Proc Natl Acad Sci USA 106: 2265–2270.
Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW . (2009b). Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature 462: 803–807.
Janz S . (2006). Myc translocations in B cell and plasma cell neoplasms. DNA Repair (Amst) 5: 1213–1224.
Janz S, Müller J, Shaughnessy J, Potter M . (1993). Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci USA 90: 7361–7365.
Kovalchuk AL, Kim JS, Janz S . (2003). E mu/S mu transposition into Myc is sometimes a precursor for T(12;15) translocation in mouse B cells. Oncogene 22: 2842–2850.
Kovalchuk AL, Kishimoto T, Janz S . (2000a). Lymph nodes and Peyer's patches of IL-6 transgenic BALB/c mice harbor T(12;15) translocated plasma cells that contain illegitimate exchanges between the immunoglobulin heavy-chain mu locus and c-myc. Leukemia 14: 1127–1135.
Kovalchuk AL, Müller JR, Janz S . (1997). Deletional remodeling of c-myc-deregulating translocations. Oncogene 15: 2369–2377.
Kovalchuk AL, Mushinski EB, Janz S . (2000b). Clonal diversification of primary BALB/c plasmacytomas harboring T(12;15) chromosomal translocations. Leukemia 14: 909–921.
Kuppers R, Dalla-Favera R . (2001). Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20: 5580–5594.
Levens DL . (2003). Reconstructing MYC. Genes Dev 17: 1071–1077.
Mitelman F, Johansson B, Mertens F . (2007). The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245.
Müller JR, Jones GM, Janz S, Potter M . (1997a). Migration of cells with immunoglobulin/c-myc recombinations in lymphoid tissues of mice. Blood 89: 291–296.
Müller JR, Jones GM, Potter M, Janz S . (1996). Detection of immunoglobulin/c-myc recombinations in mice that are resistant to plasmacytoma induction. Cancer Res 56: 419–423.
Müller JR, Mushinski EB, Jones GM, Williams JA, Janz S, Hausner PF et al. (1997b). Generation of immunoglobulin/c-myc recombinations in murine Peyer's patch follicles. Curr Top Microbiol Immunol 224: 251–255.
Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T . (2000). Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563.
Nussenzweig A, Sokol K, Burgman P, Li L, Li GC . (1997). Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci USA 94: 13588–13593.
Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C et al. (1997). Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J Exp Med 186: 921–929.
Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M et al. (2004). AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118: 431–438.
Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S et al. (2008). AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135: 1028–1038.
Silva S, Kovalchuk AL, Kim JS, Klein G, Janz S . (2003). BCL2 accelerates inflammation-induced BALB/c plasmacytomas and promotes novel tumors with coexisting T(12;15) and T(6;15) translocations. Cancer Res 63: 8656–8663.
Suematsu S, Matsusaka T, Matsuda T, Ohno S, Miyazaki J, Yamamura K et al. (1992). Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Proc Natl Acad Sci USA 89: 232–235.
Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E et al. (2009). Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 460: 231–236.
Wessels JT, Busse AC, Mahrt J, Dullin C, Grabbe E, Mueller GA . (2007). In vivo imaging in experimental preclinical tumor research—a review. Cytometry A 71: 542–549.
Willis TG, Dyer MJ . (2000). The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood 96: 808–822.
Acknowledgements
We thank our colleagues from NCI and NIAID, NIH for their contributions to this project: Tina Willington, Vaishali Jarral and Wendy DuBois for genotyping and assistance with the mouse experiments; Eileen Southon for gene targeting; Dr Alexander L Kovalchuk for advice on PCR analysis and providing primers; Drs Sung Sup Park and Santiago Silva for contributions to early stages of this project; and Drs Michael Potter and Beverly Mock for stimulating discussion and laboratory support. For assistance with transfer, cryopreservation, maintenance and rederivation of mice under SPF conditions, we thank: Ling Hu, CCOM, Iowa City, Iowa; the staff of the Jackson Laboratory, Bar Harbor, Maine; and the Office of Animal Resources, CCOM, particularly Dr Kem Singletary and James Hynes. We also thank Lorraine Tygrett, CCOM for assistance with flow cytometry. This work was supported, in part, by the Intramural Research Program of the NIH (LT, RC, TR) and Award Number P50CA097274 from the National Cancer Institute (SJ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Takizawa, M., Kim, J., Tessarollo, L. et al. Genetic reporter system for oncogenic Igh–Myc translocations in mice. Oncogene 29, 4113–4120 (2010). https://doi.org/10.1038/onc.2010.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2010.150