Abstract
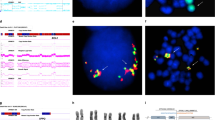
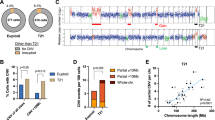
A crucial role of segmental duplications (SDs) of the human genome has been shown in chromosomal rearrangements associated with several genomic disorders. Limited knowledge is yet available on the molecular processes resulting in chromosomal rearrangements in tumors. The t(9;22)(q34;q11) rearrangement causing the 5′BCR/3′ABL gene formation has been detected in more than 90% of cases with chronic myeloid leukemia (CML). In 10–18% of patients with CML, genomic deletions were detected on der(9) chromosome next to translocation breakpoints. The molecular mechanism triggering the t(9;22) and deletions on der(9) is still speculative. Here we report a molecular cytogenetic analysis of a large series of patients with CML with der(9) deletions, revealing an evident breakpoint clustering in two regions located proximally to ABL and distally to BCR, containing an interchromosomal duplication block (SD_9/22). The deletions breakpoints distribution appeared to be strictly related to the distance from the SD_9/22. Moreover, bioinformatic analyses of the regions surrounding the SD_9/22 revealed a high Alu frequency and a poor gene density, reflecting genomic instability and susceptibility to rearrangements. On the basis of our results, we propose a three-step model for t(9;22) formation consisting of alignment of chromosomes 9 and 22 mediated by SD_9/22, spontaneous chromosome breakages and misjoining of DNA broken ends.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Albano F, Anelli L, Zagaria A, Archidiacono N, Liso V, Specchia G et al. (2007). ‘Home-brew’ FISH assay shows higher efficiency than BCR-ABL dual color, dual fusion probe in detecting microdeletions and complex rearrangements associated with t(9;22) in chronic myeloid leukemia. Cancer Genet Cytogenet 174: 121–126.
Bailey JA, Eichler EE . (2006). Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564.
Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M et al. (2004). The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet 74: 1–10.
Darai-Ramqvist E, Sandlund A, Muller S, Klein G, Imreh S, Kost-Alimova M . (2008). Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions. Genome Res 18: 370–379.
Elliott B, Jasin M . (2002). Double-strand breaks and translocations in cancer. Cell Mol Life Sci 59: 373–385.
Fourouclas N, Campbell PJ, Bench AJ, Swanton S, Baxter EJ, Huntly BJ et al. (2006). Size matters: the prognostic implications of large and small deletions of the derivative 9 chromosome in chronic myeloid leukemia. Haematologica 91: 952–955.
Gibcus JH, Kok K, Menkema L, Hermsen MA, Mastik M, Kluin PM et al. (2007). High-resolution mapping identifies a commonly amplified 11q13.3 region containing multiple genes flanked by segmental duplications. Hum Genet 121: 187–201.
Gu W, Zhang F, Lupski JR . (2008). Mechanisms for human genomic rearrangements. Pathogenetics 1: 4.
Huntly BJ, Reid AG, Bench AJ, Campbell LJ, Telford N, Shepherd P et al. (2001). Deletions of the derivative chromosome 9 occur at the time of the Philadelphia translocation and provide a powerful and independent prognostic indicator in chronic myeloid leukemia. Blood 98: 1732–1738.
Kim PM, Lam HY, Urban AE, Korbel JO, Affourtit J, Grubert F et al. (2008). Analysis of copy number variants and segmental duplications in the human genome: evidence for a change in the process of formation in recent evolutionary history. Genome Res 18: 1865–1874.
Kolomietz E, Al-Maghrabi J, Brennan S, Karaskova J, Minkin S, Lipton J et al. (2001). Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood 97: 3581–3588.
Kolomietz E, Marrano P, Yee K, Thai B, Braude I, Kolomietz A et al. (2003). Quantitative PCR identifies a minimal deleted region of 120 kb extending from the Philadelphia chromosome ABL translocation breakpoint in chronic myeloid leukemia with poor outcome. Leukemia 17: 1313–1323.
Kreil S, Pfirrmann M, Haferlach C, Waghorn K, Chase A, Hehlmann R et al. (2007). Heterogeneous prognostic impact of derivative chromosome 9 deletions in chronic myelogenous leukemia. Blood 110: 1283–1290.
Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D et al. (1990). High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247: 64–69.
Litz CE, McClure JS, Copenhaver CM, Brunning RD . (1993). Duplication of small segments within the major breakpoint cluster region in chronic myelogenous leukemia. Blood 81: 1567–1572.
Lomiento M, Jiang Z, D'Addabbo P, Eichler EE, Rocchi M . (2008). Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol 9: R173.
Mefford HC, Eichler EE . (2009). Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev 19: 196–204.
Saglio G, Storlazzi CT, Giugliano E, Surace C, Anelli L, Rege-Cambrin G et al. (2002). A 76-kb duplicon maps close to the BCR gene on chromosome 22 and the ABL gene on chromosome 9: possible involvement in the genesis of the Philadelphia chromosome translocation. Proc Natl Acad Sci USA 99: 9882–9887.
Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA et al. (2006). Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 38: 1038–1042.
Sinclair PB, Nacheva EP, Leversha M, Telford N, Chang J, Reid A et al. (2000). Large deletions at the t(9;22) breakpoint are common and may identify a poor-prognosis subgroup of patients with chronic myeloid leukemia. Blood 95: 738–743.
Specchia G, Albano F, Anelli L, Storlazzi CT, Zagaria A, Liso A et al. (2004). Derivative chromosome 9 deletions in chronic myeloid leukemia are associated with loss of tumor suppressor genes. Leuk Lymphoma 45: 689–694.
Storlazzi CT, Specchia G, Anelli L, Albano F, Pastore D, Zagaria A et al. (2002). Breakpoint characterization of der(9) deletions in chronic myeloid leukemia patients. Genes Chromosomes Cancer 35: 271–276.
Vaz de Campos MG, Montesano FT, Rodrigues MM, Chauffaille Mde L . (2007). Clinical implications of der(9q) deletions detected through dual-fusion fluorescence in situ hybridization in patients with chronic myeloid leukemia. Cancer Genet Cytogenet 178: 49–56.
Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault AC, Lupski JR . (2009). Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet 18: 1924–1936.
Acknowledgements
We thank Ms MVC Pragnell for language revision of the paper. The financial support of Associazione Italiana contro le Leucemie (AIL)-BARI is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Albano, F., Anelli, L., Zagaria, A. et al. Genomic segmental duplications on the basis of the t(9;22) rearrangement in chronic myeloid leukemia. Oncogene 29, 2509–2516 (2010). https://doi.org/10.1038/onc.2009.524
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2009.524
Keywords
This article is cited by
-
Genome-wide analysis of the GRAS gene family exhibited expansion model and functional differentiation in sea buckthorn (Hippophae rhamnoides L.)
Plant Biotechnology Reports (2021)
-
Significant Role of Segmental Duplications and SIDD Sites in Chromosomal Translocations of Hematological Malignancies: A Multi-parametric Bioinformatic Analysis
Interdisciplinary Sciences: Computational Life Sciences (2018)
-
Segmental duplications: evolution and impact among the current Lepidoptera genomes
BMC Evolutionary Biology (2017)
-
Are submicroscopic chromosomal inversions predisposing factors for the t(9;22)(q34;q11.2) translocation in chronic myeloid leukemia?
Molecular Cytogenetics (2015)
-
5‘RUNX1-3’USP42 chimeric gene in acute myeloid leukemia can occur through an insertion mechanism rather than translocation and may be mediated by genomic segmental duplications
Molecular Cytogenetics (2014)