Abstract
BACKGROUND:
Obesity is a public health concern. Yet the identification of adiposity-related genetic variants among United States (US) Hispanics, which is the largest US minority group, remains largely unknown.
OBJECTIVE:
To interrogate an a priori list of 47 (32 overall body mass and 15 central adiposity) index single-nucleotide polymorphisms (SNPs) previously studied in individuals of European descent among 3494 US Hispanic women in the Women’s Health Initiative SNP Health Association Resource (WHI SHARe).
DESIGN:
Cross-sectional analysis of measured body mass index (BMI), waist circumference (WC) and waist-to-hip ratio (WHR) were inverse normally transformed after adjusting for age, smoking, center and global ancestry. WC and WHR models were also adjusted for BMI. Genotyping was performed using the Affymetrix 6.0 array. In the absence of an a priori selected SNP, a proxy was selected (r2⩾0.8 in CEU).
RESULTS:
Six BMI loci (TMEM18, NUDT3/HMGA1, FAIM2, FTO, MC4R and KCTD15) and two WC/WHR loci (VEGFA and ITPR2-SSPN) were nominally significant (P<0.05) at the index or proxy SNP in the corresponding BMI and WC/WHR models. To account for distinct linkage disequilibrium patterns in Hispanics and further assess generalization of genetic effects at each locus, we interrogated the evidence for association at the 47 surrounding loci within 1 Mb region of the index or proxy SNP. Three additional BMI loci (FANCL, TFAP2B and ETV5) and five WC/WHR loci (DNM3-PIGC, GRB14, ADAMTS9, LY86 and MSRA) displayed Bonferroni-corrected significant associations with BMI and WC/WHR. Conditional analyses of each index SNP (or its proxy) and the most significant SNP within the 1 Mb region supported the possible presence of index-independent signals at each of these eight loci as well as at KCTD15.
CONCLUSION:
This study provides evidence for the generalization of nine BMI and seven central adiposity loci in Hispanic women. This study expands the current knowledge of common adiposity-related genetic loci to Hispanic women.
Similar content being viewed by others
Introduction
Little is known about the etiologic factors underlying the high prevalence of obesity, particularly among United States (US) minority populations. Concurrent with the obesity epidemic, US demographics have dramatically shifted. As of 2010, US Hispanics represented approximately 16% of the nation to become its largest minority group.1 Between 2009 and 2010, 41% of US Hispanic women were overweight or obese as compared with 32% of their non-Hispanic White counterparts,2 with the most notable ethnic disparities occurring among Puerto Rican and Dominican women.3 Thus, there is a rising impetus to investigate the underlying determinants of obesity among these populations.
In the past 5 years, genome-wide association studies (GWAS) have identified nearly 50 common genetic loci associated with body mass index (BMI)4, 5, 6 and anthropometric measures of central adiposity (that is, waist circumference (WC) and waist-to-hip ratio (WHR))7, 8 in European middle-aged adult populations from Europe, Australia, and the US recent GWAS in non-European ancestry populations have identified additional novel loci, including four new BMI-associated loci among East Asians, of which at least two loci do not show association in individuals of European descent3, 7, 9 and possibly three novel loci in a GWAS of BMI in individuals of African descent completed recently.9, 10, 11, 12, 13 Targeted genotyping studies of selected variants have been undertaken in Hispanic Americans.14 However, to date the contribution of genetic variants to adiposity traits in this diverse ethnic group remain largely unknown.
We investigated the associations of adiposity measures with previously identified European descent established genetic loci for BMI, WC and WHR among 3587 self-identified Hispanic women from the Women’s Health Initiative (WHI) SNP (single-nucleotide polymorphism) Health Association Resource (SHARe).
Materials and methods
WHI SHARe participants
WHI consists of multiple components including an observational study and clinical trial cohorts of postmenopausal women in the US;15 detailed recruitment and exclusion criteria have been described previously.16 Medical histories were updated annually or semi-annually by questionnaire or by phone. All participating institutions obtained Institutional Review Board approval. WHI SHARe included a total sample of 3642 self-identified Hispanic subjects from WHI, who had consented to genetic research.
Phenotypes
All phenotypic information (for example, covariate and outcome variables) was obtained during the WHI baseline questionnaires and clinic examination. Weight was measured after removing shoes, heavy clothing and pocket contents using a calibrated digital scale and recorded to the nearest one-tenth of a kilogram. Height was taken using a wall-mounted stadiometer and recorded to nearest one-tenth of a centimeter. BMI was calculated from measured height and weight (kg m–2) and was missing for 26 of the participants in our sample. WC was measured at the level of natural waist (narrowest part of torso, n=14 missing) and hips at top of the iliac crest with extra layers of clothes removed (n=13 missing) and recorded to nearest half-centimeter. WHR was then calculated as the ratio of waist to hip circumference (n=16 missing).
Genotypes
As described previously,17 DNA was extracted by the Specimen Processing Laboratory at the Fred Hutchinson Cancer Research Center (FHCRC) using white blood cells that were collected at the time of enrollment of the subjects in WHI. Specimens were stored at a central biorepository at −80 °C until analysis. Genotyping was done at Affymetrix, Inc. on the Affymetrix 6.0 array (Santa Clara, CA, USA), using 2 μg DNA at a concentration of 100 ng μl–1.
Quality control
Of the 3642 women in WHI SHARe who self-identified as Hispanic and consented for genetic testing, approximately 1% of their genetic samples could not be genotyped (n=36). We excluded samples that had call rates below 95%, which were duplicates of subjects other than their monozygotic twins, or that appeared to include a Y chromosome (that is, representing possible sample contamination, genotyping errors or an inconsistent genotypic sex; n=19). Furthermore, SNPs that were located on the Y chromosome or were Affymetrix QC probes (that is, not intended for analysis) were excluded (n=3280). SNPs with a call rate below 95% or concordance rate below 98% were flagged and excluded leaving 871 309 SNPs. These quality control measures left us with 3587 Hispanics and an average call rate of 99.8% across the 871 309 unflagged SNPs. We also excluded one person from identified relative pairs, prioritizing for complete genotype data (n=93), leading to a final analytic sample of 3494 self-identified Hispanic women.
Two hundred thirty-eight (2%) additional samples were genotyped as blind duplicates. We analyzed 188 pairs of blind duplicate samples. The overall concordance rate was 99.8% (range 95–100% over all samples, 98–100% across 871 309 SNPs that were included after genotype cleaning).
Admixture
Eigenvectors were computed in Eigenstrat18, 19 to account for global ancestry based on 178 101 markers, excluding mitochondria and sex chromosome markers, that were in common between WHI Hispanics samples and HapMap20, 21 and HGDP22 reference panels. In particular, we excluded SNPs that were A/T or C/G, on the sex chromosomes, or in the mitochondria. Individuals included from HGDP panels were 225 East Asians and 63 Native Americans, specifically 8 Surui, 22 Mayans, 13 Karitiana, 14 Pima and 6 Colombian. We also estimated proportions of European, Native American and African ancestry (Supplementary Figure 1) in the unrelated WHI SHARe sample (n=3494) using Admixture 1.22 (http://www.genetics.ucla.edu/software/admixture).
Adiposity SNP selection
One SNP from each established adiposity locus (described as of 1 July 2012 with BMI WC or WHR in GWAS of European descent individuals) was selected. A total of 47 loci were selected; 32 loci previously associated with BMI and 15 loci previously associated with WC or WHR (Tables 2a and 3a). All selected SNPs from the original publications were those that had the lowest P-value and that met genome-wide significance within a predefined locus (typically defined as 1 Mb and r2<0.1).
Generalization
We assessed generalization of previously established GWAS loci using a tiered approach. All SNPs analyzed here were originally reported in populations of European descent, so we define ‘generalization’ of a genetic effect when a SNP displays a direction of effect consistent with the original report and/or in terms of statistical significance as defined below.
First we interrogated the exact SNP from the published literature, which we defined as an ‘index SNP’. All selected index SNPs met genome-wide significance level in prior publications. To assess the consistency of effects in our study, we accessed genome-wide publically available data from the Genetic Investigation of ANthropometric Traits (GIANT) Consortium on the risk allele and its frequency in their large sample of individuals of European descent. Loci previously described with overall or central adiposity were queried in the BMI and WHR adjusted for BMI GWAS results files, respectively. If this information was missing, then we supplemented it with the relevant publication to determine directional consistency. If the previously reported adiposity SNP was not genotyped as part of WHI SHARe, the WHI SHARe SNP in highest linkage disequilibrium (LD) with the previous reported SNP (r2⩾0.8 in Hap Map CEU phase II) was selected as a proxy of the index signal. Generalization of the index or proxy SNPs was declared when directional consistency and nominal statistical significance (P<0.05) were observed.
Owing to the extensive admixture in populations of self-identified Hispanic ancestry,23, 24 we also hypothesized that even if a SNP originally identified in European or East Asian ancestry populations is not associated with BMI in those within our cohort of women who report Hispanic ancestry, the locus may still show association with a different variant in the same chromosomal region. Therefore, we searched for common variants within the established loci that better captured the association of the index SNP reported in the European and Asian populations. We identified SNPs as potentially better markers of the index signal, ‘index-dependent signals’, if they were (1) within 1 Mb of the index SNP, (2) were dependent on the index SNP in the referent population (r2⩾0.2) and (3) were associated with the anthropometric traits in our data at a significance level that was at least one order of magnitude greater than the index SNP or its proxy. In contrast, we also interrogated the evidence for possible ‘index-independent signals’ by visual inspection of all P-values of SNP–anthropometric trait associations for ‘SNPs of interest’ with r2<0.2 and within the 1 Mb region of the index SNP. Index-independent signals were deemed statistically significant if they displayed nominal significance after correcting for the total number of regions interrogated for each phenotype of interest (BMI: P=0.05/32 and WC/WHR: P=0.05/15). Conditional analyses were also conducted to confirm signal dependence. If adjustment for the index SNP decreased the P-value for the candidate index-independent signal, the SNP–phenotype association was considered suggestive evidence for an index-independent signal, without overwhelming proof that the signal was indeed independent. Certainly these signals need to be further interrogated with much larger sample sizes and/or fine mapping and within the different sub-populations of US Hispanic ancestry. In contrast, if the conditional P-value did not change or increased less than one order of magnitude in comparison with the unconditional P-value, then we declared this a possible index-independent signal. All conditional analyses were modeled in Stata 12 (StataCorp LP, College Station, TX, USA).
Statistical models
After adjusting for age, smoking status and clinical center, BMI residuals were inverse normally transformed. WC and WHR were adjusted for age, smoking status, clinical center as well as BMI, and then the residuals were inverse normally transformed. Inverse normal transformations entail creating a modified rank variable and then computing a new transformed value for the phenotype per subject such that the distribution of the phenotype is normalized with a mean of 0 and an s.d. of 1. For each of the three inverse normalized phenotypes (that is, BMI, WC adjusted for BMI and WHR adjusted for BMI) single marker linear associations further adjusted for the top 10 principal components assuming an additive model, were run using PLINK software v1.07.25 Estimated P-values below 5 × 10–8 were considered to be genome-wide significant.
LD assessment
We considered signals independent if their LD was r2<0.2 in a sample of 9345 individuals of European descent (primarily non-Hispanic Whites) from the Atherosclerosis Risk in Communities Study (ARIC). If information was not available from ARIC then HapMap CEU data (phase II or III) were used to represent the LD structure of individuals of European descent and are specifically noted in Tables 2b and 3b. In addition, estimates of LD were calculated in WHI SHARe Hispanics. LD estimates for both ARIC and WHI SHARe were calculated using the PLINK software v1.07.25
Power
We calculated estimates of power to detect associations of similar magnitude among Hispanics as those previously described in European populations across a range of common minor allele frequencies. These calculations assumed an additive genetic model, an independent sample of 3494 women, the same Bonferroni corrections and phenotype distribution as observed in our sample of US Hispanic women. Based on effect sizes published in European populations, power to detect associations was less among measures of overall (BMI) than for central adiposity (WC and WHR; Supplementary Figure 2). For example, at the minor allele frequency and previously reported effect size of FTO (32% and beta=0.39 kg m–2 change per T allele)4 we would at best have 40% statistical power to detect this effect in our study. Similarly, power to detect associations of all other BMI loci was below 80%. Moderately common WC variants (>20%) would be expected to have >80% power at mid-sized effects, which was approximately 1 cm change in WC per effect allele; whereas, most common WHR variants (>5%) frequent would be expected to have >80% power at far smaller effect sizes (approximately 0.011 WHR units). Power calculations were calculated using QUANTO v1.2.4 (http://hydra.usc.edu/gxe/).
Results
The final analytic sample of self-identified Hispanic women included in this sample was 3494. As shown in Table 1, the largest percentage of the women in this sample were between 50 and 59 years of age with a high school diploma or equivalent, were married, of Mexican ancestry, overweight in the absence of abdominal obesity as defined by the World Health Organization26, 27, 28 and were participants in a clinical trial from one of the Western or Southern WHI study centers.
Adiposity SNP generalization
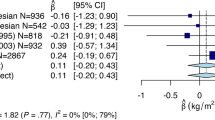
Although no SNPs reached genome-wide significance in this study, we were able to investigate the associations at 47 established obesity loci previously identified in European populations with BMI, WC and WHR in our sample of Hispanic women (Tables 2b and 3b). As summarized in Figure 1, among 16 loci with evidence of generalization 7 were defined as ‘index-dependent signals’. Of these seven, five loci were best represented by the index SNP (or its proxy) and two by a better marker in the region (defined as ‘other index-dependent signal’). A total of nine loci displayed, at least, suggestive evidence for index-independent signals as the SNP in these loci with the lowest P-value were in low LD with the index signals previously described among European descent individuals and remained nominally significant after adjustment for the index SNP (or its proxy) in conditional analyses.
Evidence for generalization of 16 previously identified obesity loci with BMI, WC and WHR in the WHI SHARe sample of Hispanic women. 1In CEU, index SNPs or proxy SNPs in LD (r2⩾0.8) below significance threshold of P<0.05. 2Identified ‘SNP of interest’ in 1 Mb region is in LD at r2⩾0.2 in CEU. P-value is Bonferroni-corrected significant and was at least one order of magnitude smaller than the P-value of the index SNP (or its proxy). 3Identified ‘SNP of interest’ in 1 Mb region is in LD at r2<0.2 in CEU. After adjustment for the index SNP (or its proxy), the P-value decreased for the ‘SNP of interest.’ 4Identified ‘SNP of interest’ in 1 Mb region is in LD at r2<0.2 in CEU. After adjustment for the index SNP (or its proxy), the P-value for the ‘SNP of interest’ did not increase more than one order of magnitude. Abbreviations: BMI, body mass index; SNP, single nucleotide polymorphism; WC, waist circumference; WHR, waist-hip ratio.
Among the 32 BMI index signals interrogated in this study, 25 had consistent directions of association as compared with publically available GIANT BMI results, which is more than expected by chance (binomial P=2.4 × 10–3). The five loci (reported above) with either evidence of generalization at the index or proxy SNP, or evidence of a better marker displayed consistent directions of effect with BMI. Among the 15 central adiposity index signals interrogated in this study, 13 had consistent directions of effects at the WC index SNP or their proxies (more than expected by chance, binomial P=3.2 × 10–3) and all had consistent directions of effect at the WHR index SNP or its proxy (binomial P=3.1 × 10–5), as compared with publically available GIANT WHR adjusted for BMI results. WC/WHR loci with either evidence of generalization at the index or proxy SNP, or evidence of a better marker displayed consistent direction of effects with the central adiposity phenotype, for which they were previously reported.
Among index or proxy SNPs selected, the BMI phenotype showed the strongest association with rs9939609 in the FTO locus (beta (s.e.)=0.085 (0.026); P=0.001), followed rs11084753 in the KCT615 locus (beta (s.e.)=−0.070 (0.025); P=0.006). Other nominally significant (P<0.05) loci were found for MC4R (rs571312), NUDT3/HMGA1 (rs378560), FAIM2 (rs7139903) and TMEM18 (rs6548238). Again, among the index or proxy SNPs selected, the strongest association with WC and WHR was found with rs60905288 near VEGFA (WC: beta (s.e.)=−0.075 (0.024); WHR: beta (s.e.)=−0.072 (0.024); P=0.002 for both). A locus near ITPR2-SSPN showed a nominal association with WHR (rs12814794, beta (s.e.)=0.050 (0.025); P=0.04).
For 11 adiposity loci (6 BMI and 5 WC/WHR), we observed a ‘SNP of interest’ with at least one order of magnitude smaller P-value than the index SNP or its proxy. These loci are displayed in Supplementary Figures 3–6. SNPs at two loci previously associated with BMI, NUDT3/HMGA1 (rs6925243, P=7.84 × 10–4), and MC4R (rs1942867; P=1.95 × 10–5) were dependent on their respective index or proxy SNPs in CEU (r2⩾0.2), and therefore were considered to represent a better marker for Hispanics at the index signal (Table 2b, Supplementary Figures 6a and b).
Four BMI loci (FANCL, ETV5, TFAP2B and KCTD15; Supplementary Figures 3a–d), and five WC or WHR loci (DNM3-PIGC, GRB14, ADAMTS9, LY86 and MSRA; Supplementary Figures 4a–d and 5a–c) had low LD in HapMap CEU populations (r2<0.2; Tables 2b and 3b) and were therefore considered as possible index-independent signals. All conditional P-values for the ‘SNP of interest’–phenotype association remained nominally significant after adjustment for the index SNP or its proxy (P<0.05). One BMI locus (TFAP2B) appeared to have suggestive evidence of an index-independent signal as the P-value decreased from the unconditional analysis for the association between the ‘SNP of interest’ and BMI (Table 4). However, the evidence for association at three BMI loci (near FANCL, ETV5 and KCTD15) for the ‘SNP of interest’–BMI association became weaker on adjustment for the index SNP or its proxy. KCTD15 was the only locus of these nine loci to have significant evidence of both generalization at both the index signal (P=0.006) and an independent signal (r2<0.2; Tables 2a and 2b). At two central adiposity loci (near DNM3-PIGC and MSRA), there was suggestive evidence of index-independent signals for WC (Table 4). In contrast, at three previously described WHR loci (GRB14, ADAMTS9 and LY86) there was inconsistent evidence across the central adiposity phenotypes tested (WC and WHR models adjusted for BMI).
Discussion
In this study of postmenopausal Hispanic women, we found that the majority of the 47 SNPs interrogated showed consistent direction of effect. Specifically, 25 of 32 SNPs for BMI (binomial test: P<7.8E–04), and 13 and 15 of 15 SNPs for WC and WHR (binomial test: P<3.2E–03 and P<3.1E–05), respectively. Further, we found associations of nine loci with BMI and seven loci with waist phenotypes (WC or WHR) previously shown to be associated with these traits in European populations from Europe, Australia and the United States. In addition, we present possible evidence for independent signals among Hispanics at nine of these loci, three of which became stronger after conditioning (locus near TFAP2B with BMI, and loci near DNM3-PIGC and MSRA with WC or WHR). As verification for the associations of these possible independent signals identified in the Hispanic women in samples of European descent individuals, we looked up the P-value in the original published results (Heid and Speliotes references) for seven of the nine SNPs that were available. None of the seven SNPs were even nominally significant (all P>0.05).
Certainly the analyses conducted here needs to be independently verified in an additional Hispanic ancestry sample. Although the exact functional variants underlying these signals still remain to be identified, it is interesting to note that TFAP2B encodes a transcription factor that has previously been associated with both BMI and type 2 diabetes in primarily non-Hispanic populations.29, 30, 31 MSRA encodes a protein that is thought to repair of oxidative damage to proteins to restore biological activity.32 Deletion of this gene has been associated with insulin resistance in mice.33 Dynamin 3 (DNM3), a member of the dynamin family of enzymes, and phosphatidylinositol glycan anchor biosynthesis, class C (PIGC), are involved in cell membrane interactions and adhesion of proteins to the cell membrane.34, 35, 36 Functional roles of some of the loci with possibly independent signals among Hispanics may include energy homeostasis for KCTD15 and ETV5 loci that are highly expressed in the hypothalamus,37 and insulin signaling from ADAMTS9, and GRB14 loci, particularly in muscle tissue.38, 39, 40, 41
Although the possibility of multiple signals at established GWAS loci needs to be confirmed in additional, larger samples of Hispanic ancestry, these findings add to the growing literature that indicates multiple variants for BMI, lipids and other complex traits. Moreover, these study findings also add to the growing literature that demonstrates suggestive generalizability of genetic loci across ancestrally distinct populations for some but not all loci. For example, for the SNPs associated with BMI in our Hispanic population, using the index SNP (or proxy SNP in LD, r2>0.5) identified in European descent populations, six loci (TFAP2B, ETV5, TMEM18, FAIM2, FTO and MC4R) also displayed directionally consistent and statistically significant associations with BMI in two large GWAS studies of BMI in African Americans and Asians.11, 13 In addition, two other BMI loci displayed directionally consistent effect estimates for BMI in Hispanics (NUDT3 and KCTD15), but did not display statistical significance. These study findings demonstrate, for the first time, a general relevance of these BMI loci across multiple ancestrally diverse US minority populations. Although we provide some evidence for generalization, our sample size is small and further verification of these findings is necessary. Further, of note, our data demonstrate often substantial differences in allele frequencies between the reference HapMap CEU population and the female participants form the WHI SHARE study. Although this study only summarizes data from a single group of Hispanics, 43% with Mexican origins and, on average, 33% Native American ancestry (Supplementary Figure 1), these data do demonstrate the extensive diversity of the Hispanic population and the critical need for a greater focus on the genetic architecture in ethnic minority populations.
It is of interest to note that FANCL did not display any evidence for generalization in African and Asian ancestry populations. In Hispanics, we detected evidence for the proxy SNP and also for a possible independent signal, suggesting distinctions at this locus across ancestral populations.
There have been fewer genetic epidemiological studies of WC and WHR in ancestrally diverse populations—perhaps because waist traits are collected less frequently in large cohorts. One study in a sample of South Asian descent found that SNPs in LD with the identified index SNP (rs1095252) near the GRB14 loci were associated with WHR and type 2 diabetes.42 In Hispanic women, we identified a possible index-dependent signal at this locus supporting the relevance of this locus across populations.
In this study, we were able to generalize or find evidence of association at 9 of 32 BMI loci (28%) and 7 of 15 WC/WHR loci (47%). Even among those loci that did not generalize in this study, the majority exhibited consistent directions of effect. The greater proportion of findings at central adiposity loci may likely be due to the greater power to detect associations. As shown in Supplementary Figures 2a–c, power calculations revealed disparate curves for overall (BMI) versus central adiposity measures (WC or WHR), wherein we were underpowered (<80% power) to detect effects for BMI. Between WC and WHR, we were most powered to detect effects on WHR. Of note, these calculations were based on a range of allele frequencies and effect sizes, as well as the distribution of the phenotype in WHI SHARe. Among the three measurements, BMI had the highest level of variability (z-score=5.2), followed by WC (z-score=7.0) and WHR (z-score=11.7), respectively. Similarly, the BMI findings were subjected to a higher penalty of Bonferroni correction (that is, lower alpha), because of the greater number of variants tested. We may also have had greater power to detect waist-related traits as WHI comprises women only, and we have recently established a stronger magnitude of genetic effects in women for many of the established waist variants.43
National estimates from 1982 to 1984 from the Hispanic Health and Nutritional Examination Survey44 were among the first to show that the burden of obesity may not be similar across all adult US Hispanics. More recent data from a diverse cohort study of four US communities strongly supports the possibility that there may be disparities in obesity among this ethic group by country of origin, with Puerto Rican women have the highest prevalence of obesity (51%) followed closely by Dominican (42%), Central American (42%) and Mexican women (42%).3 Unfortunately, although the WHI Hispanic sample included in WHI SHARe roughly corresponds to the distribution of US Hispanics recorded in the 2010 Census (63% of Hispanics of Mexican descent),30 it likely does not capture the true diversity that constitutes this ethnicity nor does it imply that these results can be generalized to all US Hispanics. Moreover, the small sample size limited our ability to assess heterogeneity of effect size by population of origin. Future research should be designed and powered to investigate genetic effects across diverse Hispanic backgrounds.
Finally, our sample of primarily postmenopausal women may have been a limitation as there is evidence that genetic effects on adiposity vary substantially across the life course.45, 46, 47
In turn, this study is strengthened by a number of factors. First, obesity-related racial/ethnic- and gender disparities exist among the largest US minority group—Hispanics48, 49, 50, 51 and progress in the obesity field will only be made when all US populations are successfully interrogated. Therefore, our interrogation of established BMI and WC/WHR loci in an ancestrally diverse population with heightened disparities in disease risk is timely and of great public health significance. Second, to our knowledge this study constitutes the first attempt in the scientific literature to perform a large comprehensive study of multiple adiposity phenotypes among a sample of Hispanic individuals. Although previous generalization studies have been published among Hispanics, they were largely conducted in the context of candidate gene studies and did not evaluate well established GWAS variants.
In summary, our findings suggest similar genetic influences on body size and shape across non-Hispanic and Hispanic descent populations, by illustrating associations at nine BMI loci and seven WC/WHR loci previously reported in European descent populations. We also provide tentative evidence that several of the BMI and WC loci harbor multiple independent signals, which has been shown to increase the heritability explained for complex traits across populations. Nonetheless, replication of these signals in larger Hispanic studies is required, as well as GWAS studies to determine if novel obesity loci can be mapped in Hispanic populations.
References
Ennis SR, Ríos-Vargas M, Albert NG . The Hispanic Population: 2010, in 2010 Census Briefs, US Census, Editor 2011. US Dept. of Commerce: Washington, DC, USA.
Flegal KM, Carroll MD, Kit BK, Ogden CL . Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA 2012; 307: 491–497.
Daviglus ML, Talavera GA, Aviles-Santa ML, Allison M, Cai J, Criqui MH et al. Prevalence of major cardiovascular risk factors and cardiovascular diseases among Hispanic/Latino individuals of diverse backgrounds in the United States. JAMA 2012; 308: 1775–1784.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249 796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Thorleifsson G, Walters G, Gudbjartsson D, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24.
Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 41: 25–34.
Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 2010; 42: 949–960.
Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet 2009; 5: e1000508.
Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet 2012; 44: 302–306.
Wang J, Mei H, Chen W, Jiang Y, Sun W, Li F et al. Study of eight GWAS-identified common variants for association with obesity-related indices in Chinese children at puberty. Int J Obes (Lond) 2012; 36: 542–547.
Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet 2012; 44: 307–311.
Ng MC, Hester JM, Wing MR, Li J, Xu J, Hicks PJ et al. Genome-wide association of BMI in African Americans. Obesity (Silver Spring) 2012; 20: 622–627.
Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet 2013; 45: 690–696.
Wing MR, Ziegler J, Langefeld CD, Ng MC, Haffner SM, Norris JM et al. Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS Family Study. Hum Genet 2009; 125: 615–626.
Anderson G, Cummings S, Freedman LS, Furberg C, Henderson M, Johnson SR et al. Design of the Women's Health Initiative Clinical Trial and Observational Study. Controlled Clinical Trials 1998; 19: 61–109.
Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C et al. The Women's Health Initiative recruitment methods and results. Ann Epidemiol 2003; 13 (9 Suppl): S18–S77.
Chen CT, Fernandez-Rhodes L, Brzyski RG, Carlson CS, Chen Z, Heiss G et al. Replication of loci influencing ages at menarche and menopause in Hispanic women: the Women's Health Initiative SHARe Study. Hum Mol Genet 2012; 21: 1419–1432.
Patterson N, Price AL, Reich D . Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
International HapMap, C. The International HapMap Project. Nature 2003; 426: 789–796.
Thorisson GA, Smith AV, Krishnan L, Stein LD . The International HapMap Project Web site. Genome Res 2005; 15: 1592–1593.
Cavalli-Sforza LL . The Human Genome Diversity Project: past, present and future. Nat Rev Genet 2005; 6: 333–340.
Buchanan CC, Torstenson ES, Bush WS, Ritchie MD . A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. J Am Med Inform Assoc 2012; 19: 289–294.
Palmer ND, Lehtinen AB, Langefeld CD, Campbell JK, Haffner SM, Norris JM et al. Association of TCF7L2 gene polymorphisms with reduced acute insulin response in Hispanic Americans. J Clin Endocrinol Metab 2008; 93: 304–309.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Committee, WE. Physical status: the use and interpretation of anthropometry. Report of a WHO Expert Committee. World Health Organ Tech Rep Ser 1995; 854: 1–452.
Consultation, WE. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 2004; 363: 157–163.
World Health Organization (WHO). Waist circumference and waist-hip ratio: report of a WHO expert consultation, Geneva, 8–11 December 2008. WHO: Geneva, Switzerland, 2011.
Maeda S, Tsukada S, Kanazawa A, Sekine A, Tsunoda T, Koya D et al. Genetic variations in the gene encoding TFAP2B are associated with type 2 diabetes mellitus. J Hum Genet 2005; 50: 283–292.
Tsukada S, Tanaka Y, Maegawa H, Kashiwagi A, Kawamori R, Maeda S . Intronic polymorphisms within TFAP2B regulate transcriptional activity and affect adipocytokine gene expression in differentiated adipocytes. Mol Endocrinol 2006; 20: 1104–1111.
Yeung E, Qi L, Hu FB, Zhang C . Novel abdominal adiposity genes and the risk of type 2 diabetes: findings from two prospective cohorts. Int J Mol Epidemiol Genet 2011; 2: 138–144.
NCBI. Entrez gene: MSRA methionine sulfoxide reductase A, 2012.
Styskal J, Nwagwu FA, Watkins YN, Liang H, Richardson A, Musi N et al. Methionine sulfoxide reductase A affects insulin resistance by protecting insulin receptor function. Free Radic Biol Med 2012; 56: 123–132.
Inoue N, Watanabe R, Takeda J, Kinoshita T . PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem Biophys Res Commun 1996; 226: 193–199.
Orth JB, McNiven MA . Dynamin at the actin-membrane interface. Curr Opin Cell Biol 2003; 15: 31–39.
Watanabe R, Inoue N, Westfall B, Taron CH, Orlean P, Takeda J et al. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J 1998; 17: 877–885.
Schmid PM, Heid I, Buechler C, Steege A, Resch M, Birner C et al. Expression of fourteen novel obesity-related genes in Zucker diabetic fatty rats. Cardiovasc Diabetol 2012; 11: 48.
Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–645.
Boesgaard TW, Gjesing AP, Grarup N, Rutanen J, Jansson PA, Hribal ML et al. Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients–EUGENE2 study. PLoS One 2009; 4: e7236.
Depetris RS, Hu J, Gimpelevich I, Holt LJ, Daly RJ, Hubbard SR . Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell 2005; 20: 325–333.
Holt LJ, Siddle K . Grb10 and Grb14: enigmatic regulators of insulin action--and more? Biochem J 2005; 388 (Pt 2): 393–406.
Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 2011; 43: 984–989.
Randall J, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL . Sex-stratified genome-wide association studies including 270 000 individuals show sexual dimorphism in genetic loci for anthropometric traits. Plos Genet 2013; 9: e1003500.
Aponte J . Diabetes-related risk factors across Hispanic subgroups in the Hispanic health and nutritional examination survey (1982-1984). Public Health Nurs 2009; 26: 23–38.
Graff M, North KE, Mohlke KL, Lange LA, Luo J, Harris KM et al. Estimation of genetic effects on BMI during adolescence in an ethnically diverse cohort: the National Longitudinal Study of Adolescent Health. Nutri Diabetes 2012; 2: e47.
Hallman DM, Friedel VC, Eissa MA, Boerwinkle E, Huber JC Jr, Harrist RB et al. The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes (Lond) 2012; 36: 61–68.
Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet 2010; 19: 545–552.
Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM . Prevalence of overweight and obesity in the United States, 1999-2004. JAMA 2006; 295: 1549–1555.
Ogden CL, Lamb MM, Carroll MD, Flegal KM . Obesity and socioeconomic status in adults: United States, 2005-2008. NCHS Data Brief 2010; 1–8.
Flegal KM, Ezzati TM, Harris MI, Haynes SG, Juarez RZ, Knowler WC et al. Prevalence of diabetes in Mexican Americans, Cubans, and Puerto Ricans from the Hispanic Health and Nutrition Examination Survey, 1982-1984. Diabetes Care 1991; 14: 628–638.
Flegal KM, Ogden CL, Yanovski JA, Freedman DS, Shepherd JA, Graubard BI et al. High adiposity and high body mass index-for-age in US children and adolescents overall and by race-ethnic group. Am J Clin Nutr 2010; 91: 1020–1026.
Acknowledgements
This work was supported by the National Heart, Lung and Blood Institute of the National Institutes of Health, US Department of Health and Human Services (grants 5T32HL007055 to LF-R, 5K07CA136969 to HMO-B). WHI SHARe was also supported by the National Heart, Lung and Blood Institute through the following contracts: N01WH22110, 24152, 32100–2, 32105–6, 32108–9, 32111–13, 32115, 32118–9, 32122, 42107–26, 42129–32 and 44221. WHI has approved the manuscript, which was prepared in collaboration with WHI investigators.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
MG, LF-R, KM and KEN defined the study approach and aims; MG, LF-R and KM completed the data analysis; MG, LF-R and KEN contributed to the interpretation and presentation of the study findings; all other authors reviewed the manuscript and provided their critical feedback and approval.
Supplementary Information accompanies this paper on the Nutrition & Diabetes website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Graff, M., Fernández-Rhodes, L., Liu, S. et al. Generalization of adiposity genetic loci to US Hispanic women. Nutr & Diabetes 3, e85 (2013). https://doi.org/10.1038/nutd.2013.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nutd.2013.26
Keywords
This article is cited by
-
Impact of Amerind ancestry and FADS genetic variation on omega-3 deficiency and cardiometabolic traits in Hispanic populations
Communications Biology (2021)
-
Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci
Human Genetics (2017)
-
Interaction of smoking and obesity susceptibility loci on adolescent BMI: The National Longitudinal Study of Adolescent to Adult Health
BMC Genetics (2015)