Abstract
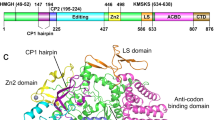
Leucyl-tRNA synthetase (LeuRS) has a specific post-transfer editing activity directed against mischarged isoleucine and similar noncognate amino acids. We describe the post-transfer–editing and product complexes of Thermus thermophilus LeuRS (LeuRSTT) with tRNALeu at 2.9- to 3.3-Å resolution. In the post-transfer–editing configuration, A76 binds in the editing active site exactly as previously found for the adenosine moiety of a small-molecule editing-substrate analog. The 60 C-terminal residues of LeuRSTT, unseen in previous structures, fold into a compact domain flexibly linked to the rest of the molecule and interacting with the G19-C56 tertiary base pair of tRNALeu. LeuRS recognition of tRNALeu depends essentially on tRNA shape rather than base-specific interactions. The structures show that considerable domain rotations, notably of the editing domain, accompany the tRNA–3′ end dynamics associated successively with aminoacylation, post-transfer editing and product release.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pauling, L. The probability of errors in the process of synthesis of protein molecules. in Arbeiten aus dem Gebiet der Naturstoffe (Festschrift Prof. Dr. Arthur Stoll) 597–602 (Birkhauser Verlag, Basel, Switzerland, 1957).
Jakubowski, H. Accuracy of aminoacyl-tRNA synthetases: proofreading of amino acids. in Aminoacyl-tRNA Synthetases (eds. Ibba, M., Francklyn, C. and Cusack, S.) 384–396 (Landes Biosciences, Georgetown, Texas, USA, 2005).
Lincecum, T.L., Jr . et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell 11, 951–963 (2003).
Fukunaga, R., Fukai, S., Ishitani, R., Nureki, O. & Yokoyama, S. Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine. J. Biol. Chem. 279, 8396–8402 (2004).
Fukai, S. et al. Structural basis for double-sieve discrimination of L-valine from L- isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell 103, 793–803 (2000).
Dock-Bregeon, A. et al. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell 103, 877–884 (2000).
Wong, F.C., Beuning, P.J., Nagan, M., Shiba, K. & Musier-Forsyth, K. Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry 41, 7108–7115 (2002).
Beebe, K., Ribas De Pouplana, L. & Schimmel, P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22, 668–675 (2003).
Roy, H., Ling, J., Irnov, M. & Ibba, M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648 (2004).
Dock-Bregeon, A.C. et al. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell 16, 375–386 (2004).
Biou, V., Yaremchuk, A., Tukalo, M. & Cusack, S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science 263, 1404–1410 (1994).
Yaremchuk, A., Kriklivyi, I., Tukalo, M. & Cusack, S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 21, 3829–3840 (2002).
Asahara, H. et al. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol. 231, 219–229 (1993).
Soma, A., Uchiyama, K., Sakamoto, T., Maeda, M. & Himeno, H. Unique recognition style of tRNA(Leu) by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 293, 1029–1038 (1999).
Asahara, H., Nameki, N. & Hasegawa, T. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol. 283, 605–618 (1998).
Tocchini-Valentini, G., Saks, M.E. & Abelson, J. tRNA leucine identity and recognition sets. J. Mol. Biol. 298, 779–793 (2000).
Larkin, D.C., Williams, A.M., Martinis, S.A. & Fox, G.E. Identification of essential domains for Escherichia coli tRNA(leu) aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 30, 2103–2113 (2002).
Du, X. & Wang, E.D. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 31, 2865–2872 (2003).
Cusack, S., Yaremchuk, A. & Tukalo, M. The crystal structure of the ternary complex of T.thermophilus seryl- tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 15, 2834–2842 (1996).
Fukunaga, R. & Yokoyama, S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol., advance online publication 11 September 2005 (10.1038/nsmb985)
Cusack, S., Yaremchuk, A. & Tukalo, M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 (2000).
Hauenstein, S., Zhang, C.M., Hou, Y.M. & Perona, J.J. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 11, 1134–1141 (2004).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Fukunaga, R. & Yokoyama, S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 346, 57–71 (2005).
Delagoutte, B., Moras, D. & Cavarelli, J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 19, 5599–5610 (2000).
Silvian, L.F., Wang, J. & Steitz, T.A. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science 285, 1074–1077 (1999).
Yaremchuk, A., Cusack, S., Gudzera, O., Grotli, M. & Tukalo, M. Crystallization and preliminary crystallographic analysis of Thermus thermophilus leucyl-tRNA synthetase and its complexes with leucine and a non-hydrolysable leucyl-adenylate analogue. Acta Crystallogr. D Biol. Crystallogr. 56, 667–669 (2000).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47, 110–119 (1991).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Esnouf, R.M. Further additions to Molscript version 1.4, including reading and contouring of electron density maps. Acta Crystallogr. D Biol. Crystallogr. 55, 938–940 (1999).
Merritt, E.A. & Murphy, M.E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50, 869–873 (1994).
Acknowledgements
The authors thank the European Synchrotron Radiation Facility (ESRF)–European Molecular Biology Laboratory Joint Structural Biology Group for access to ESRF synchrotron beamline facilities. A.Y. was supported in part by the Human Frontiers Science Programme Research grant RGP0190/2001-M and M.T. by US National Institutes of Health grant GM63107 to S. Martinis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Multiple sequence alignment of the C-terminal domain of representative bacteria and organellar LeuRS showing conservation of key residues. (PDF 68 kb)
Supplementary Fig. 2
Simulated omit map Fo-Fc electron density for the 3′ end of tRNALeu and Nva2AA. (PDF 302 kb)
Rights and permissions
About this article
Cite this article
Tukalo, M., Yaremchuk, A., Fukunaga, R. et al. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer–editing conformation. Nat Struct Mol Biol 12, 923–930 (2005). https://doi.org/10.1038/nsmb986
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb986
This article is cited by
-
Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip
Communications Biology (2022)
-
Deciphering the interaction of benzoxaborole inhibitor AN2690 with connective polypeptide 1 (CP1) editing domain of Leishmania donovani leucyl-tRNA synthetase
Journal of Biosciences (2020)
-
ECOD: identification of distant homology among multidomain and transmembrane domain proteins
BMC Molecular and Cell Biology (2019)
-
Structural characterization of antibiotic self-immunity tRNA synthetase in plant tumour biocontrol agent
Nature Communications (2016)
-
Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling
Cell Discovery (2016)