Abstract
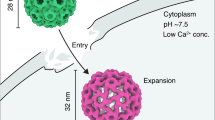
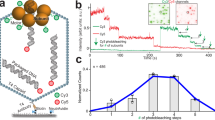
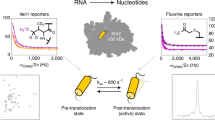
Molecular motors undergo cyclical conformational changes and convert chemical energy into mechanical work. The conformational dynamics of a viral packaging motor, the hexameric helicase P4 of dsRNA bacteriophage φ8, was visualized by hydrogen-deuterium exchange and high-resolution mass spectrometry. Concerted changes of exchange kinetics revealed a cooperative unit that dynamically links ATP-binding sites and the central RNA-binding channel. The cooperative unit is compatible with a structure-based model in which translocation is mediated by a swiveling helix. Deuterium labeling also revealed the transition state associated with RNA loading, which proceeds via opening of the hexameric ring. The loading mechanism is similar to that of other hexameric helicases. Hydrogen-deuterium exchange provides an important link between time-resolved spectroscopic observations and high-resolution structural snapshots of molecular machines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Keller, D. & Bustamante, C. The mechanochemistry of molecular motors. Biophys. J. 78, 541–556 (2000).
Delagoutte, E. & von Hippel, P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Quart. Rev. Biophys. 35, 431–478 (2002).
Moore, S.D. & Prevelige, P.E. Jr. DNA packaging: a new class of molecular motors. Curr. Biol. 12, R96–R98 (2002).
Mancini, E.J. et al. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118, 743–755 (2004).
Kainov, D.E. et al. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J. Biol. Chem. 278, 48084–48091 (2003).
Singleton, M.R., Sawaya, M.R., Ellenberger, T. & Wigley, D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000).
Patel, S.S. & Picha, K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000).
Skordalakes, E. & Berger, J.M. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003).
Gai, D., Zhao, R., Li, D., Finkielstein, C.V. & Chen, X.S. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 (2004).
Hoofnagle, A.N., Resing, K.A. & Ahn, N.G. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 32, 1–25 (2003).
Englander, S.W., Sosnick, T.R., Englander, J.J. & Mayne, L. Mechanisms and uses of hydrogen exchange. Curr. Opin. Struct. Biol. 6, 18–23 (1996).
Hoofnagle, A.N., Resing, K.A., Goldsmith, E.J. & Ahn, N.G. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc. Natl. Acad. Sci. USA 98, 956–961 (2001).
Lanman, J. et al. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11, 676–677 (2004).
Marshall, A.G., Hendrickson, C.L. & Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 17, 1–35 (1998).
Wang, F. et al. Fourier transform ion cyclotron resonance mass spectrometric detection of small Ca(2+)-induced conformational changes in the regulatory domain of human cardiac troponin C. J. Am. Soc. Mass Spectrom. 10, 703–710 (1999).
Lisal, J., Kainov, D.E., Bamford, D.H., Thomas, G.J. Jr. & Tuma, R. Enzymatic mechanism of RNA translocation in double-stranded RNA bacteriophages. J. Biol. Chem. 279, 1343–1350 (2004).
Milne, J.S., Mayne, L., Roder, H., Wand, A.J. & Englander, S.W. Determinants of protein hydrogen exchange studied in equine cytochrome c. Protein Sci. 7, 739–745 (1998).
Englander, J.J. et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA 100, 7057–7062 (2003).
Picha, K.M., Ahnert, P. & Patel, S.S. DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process. Nucleotide hydrolysis is not required. Biochemistry 39, 6401–6409 (2000).
Zhang, Z., Post, C.B. & Smith, D.L. Amide hydrogen exchange determined by mass spectrometry: application to rabbit muscle aldolase. Biochemistry 35, 779–791 (1996).
Frilander, M. & Bamford, D.H. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage φ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 246, 418–428 (1995).
Kim, D.E. & Patel, S.S. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J. Biol. Chem. 276, 13902–13910 (2001).
Ahnert, P., Picha, K.M. & Patel, S.S. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 19, 3418–3427 (2000).
Davey, M.J., Jeruzalmi, D., Kuriyan, J. & O'Donnell, M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3, 826–835 (2002).
Mindich, L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage φ6. Microbiol. Mol. Biol. Rev. 63, 149–160 (1999).
Kainov, D.E., Lisal, J., Bamford, D.H. & Tuma, R. Packaging motor from double-stranded RNA bacteriophage φ12 acts as an obligatory passive conduit during transcription. Nucleic Acids Res. 32, 3515–3521 (2004).
Juuti, J.T., Bamford, D.H., Tuma, R. & Thomas, G.J., Jr. Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ6. J. Mol. Biol. 279, 347–359 (1998).
Wooll, J.O., Wrabl, J.O. & Hilser, V.J. Ensemble modulation as an origin of denaturant-independent hydrogen exchange in proteins. J. Mol. Biol. 301, 247–256 (2000).
Hilser, V.J. & Freire, E. Structure-based calculation of the equilibrium folding pathway of proteins. Correlation with hydrogen exchange protection factors. J. Mol. Biol. 262, 756–772 (1996).
Hilser, V.J. Modeling the native state ensemble. Methods Mol. Biol. 168, 93–116 (2001).
Kainov, D.E., Butcher, S.J., Bamford, D.H. & Tuma, R. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J. Mol. Biol. 328, 791–804 (2003).
Lam, T.T. et al. Mapping of protein:protein contact surfaces by hydrogen/deuterium exchange, followed by on-line high-performance liquid chromatography-electrospray ionization Fourier-transform ion-cyclotron-resonance mass analysis. J. Chromatogr. A 982, 85–95 (2002).
Emmett, M.R. & Caprioli, R.M. Micro-electrospray mass spectrometry: ultra-high-sensitivity analysis of peptides and proteins. J. Am. Soc. Mass Spectrom. 5, 605–613 (1994).
Emmett, M.R., White, F.M., Hendrickson, C.L., Shi, S.D.-H. & Marshall, A.G. Application of micro-electrospray liquid chromatographic techniques to FT-ICR MS to enable high-sensitivity biological analysis. J. Am. Soc. Mass Spectrom. 9, 333–340 (1998).
Senko, M.W., Hendrickson, C.L., Emmett, M.R., Shi, S.D.-H. & Marshall, A.G. External accumulation of ions for enhanced electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 8, 970–976 (1997).
Zhang, Z., Li, W., Logan, T.M., Li, M. & Marshall, A.G. Human recombinant [C22A] FK506-binding protein amide hydrogen exchange rates from mass spectrometry match and extend those from NMR. Protein Sci. 6, 2203–2217 (1997).
Huang, C.C., Couch, G.S., Pettersen, E.F. & Ferrin, T.E. Chimera: an extensible molecular modeling application constructed using standard components. Pac. Symp. Biocomput. 1, 724–725 (1996).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Acknowledgements
We thank A. Abu Ramadan for the software development. J.L. is supported by Viikki Graduate School in Biosciences, D.E.K is supported by the National Graduate School in Informational and Structural Biology. This work was supported by Academy of Finland grant 206926 (R.T.), the Finnish Centre of Excellence Program 2000–2005, the US National Science Foundation National High Field FT-ICR Mass Spectrometry Facility (CHE-94-13008), Florida State University and the National High Magnetic Field Laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Sequence alignment. (PDF 1989 kb)
Supplementary Fig. 2
Bimodal isotopic distributions during RNA loading. (PDF 50 kb)
Rights and permissions
About this article
Cite this article
Lísal, J., Lam, T., Kainov, D. et al. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat Struct Mol Biol 12, 460–466 (2005). https://doi.org/10.1038/nsmb927
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb927
This article is cited by
-
Rapid Screening for Potential Epitopes Reactive with a Polycolonal Antibody by Solution-Phase H/D Exchange Monitored by FT-ICR Mass Spectrometry
Journal of the American Society for Mass Spectrometry (2013)
-
Intersubunit coordination in a homomeric ring ATPase
Nature (2009)