Abstract
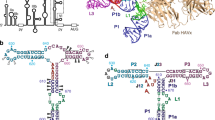
The cocrystal structure of the PP7 bacteriophage coat protein in complex with its translational operator identifies a distinct mode of sequence-specific RNA recognition when compared to the well-characterized MS2 coat protein–RNA complex. The structure reveals the molecular basis of the PP7 coat protein's ability to selectively bind its cognate RNA, and it demonstrates that the conserved β-sheet surface is a flexible architecture that can evolve to recognize diverse RNA hairpins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kozak, M. & Nathans, D. Bacteriol. Rev. 36, 109–134 (1972).
Olsthoorn, R.C., Garde, G., Dayhuff, T., Atkins, J.F. & Van Duin, J. Virology 206, 611–625 (1995).
Carey, J., Cameron, V., de Haseth, P.L. & Uhlenbeck, O.C. Biochemistry 22, 2601–2610 (1983).
Lim, F., Downey, T.P. & Peabody, D.S. J. Biol. Chem. 276, 22507–22513 (2001).
Spingola, M. & Peabody, D.S. Nucleic Acids Res. 25, 2808–2815 (1997).
Lim, F., Spingola, M. & Peabody, D.S. J. Biol. Chem. 271, 31839–31845 (1996).
Horn, W.T. et al. Structure 14, 487–495 (2006).
Valegard, K., Liljas, L., Fridborg, K. & Unge, T. Nature 345, 36–41 (1990).
Ni, C.Z. et al. Structure 3, 255–263 (1995).
Golmohammadi, R., Fridborg, K., Bundule, M., Valegard, K. & Liljas, L. Structure 4, 543–554 (1996).
Tars, K., Bundule, M., Fridborg, K. & Liljas, L. J. Mol. Biol. 271, 759–773 (1997).
Lim, F. & Peabody, D.S. Nucleic Acids Res. 30, 4138–4144 (2002).
Valegard, K. et al. J. Mol. Biol. 270, 724–738 (1997).
Wu, H.N. & Uhlenbeck, O.C. Biochemistry 26, 8221–8227 (1987).
Valegard, K., Murray, J.B., Stockley, P.G., Stonehouse, N.J. & Liljas, L. Nature 371, 623–626 (1994).
Bardwell, V.J. & Wickens, M. Nucleic Acids Res. 18, 6587–6594 (1990).
Hogg, J.R. & Collins, K. RNA 13, 868–880 (2007).
Lykke-Andersen, J., Shu, M.D. & Steitz, J.A. Cell 103, 1121–1131 (2000).
Bertrand, E. et al. Mol. Cell 2, 437–445 (1998).
Acknowledgements
This work was supported by the US National Institutes of Health (grants AR-41480 and EB-002060 to R.H.S and National Research Service Award institutional training grant (5T32HL007675-19) support to J.A.C.) and the Albert Einstein Cancer Center. The authors wish to thank the staff at the National Synchrotron Light Source X29a beamline for assistance with data collection, the staff at the Argonne Advanced Photon Source Structural GenomiX Collaborative Access Team beamline for express crystallography data collection and G. Arenas, M. Hennig, U. Meier, S. Nguyen and S. Ryder for helpful discussions.
Author information
Authors and Affiliations
Contributions
J.A.C. designed and performed the experiments. Y.P. assisted with crystallography. J.A.C., Y.P., S.C.A. and R.H.S. wrote and discussed the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 and Supplementary Methods (PDF 5810 kb)
Rights and permissions
About this article
Cite this article
Chao, J., Patskovsky, Y., Almo, S. et al. Structural basis for the coevolution of a viral RNA–protein complex. Nat Struct Mol Biol 15, 103–105 (2008). https://doi.org/10.1038/nsmb1327
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1327
This article is cited by
-
Antiviral responses are shaped by heterogeneity in viral replication dynamics
Nature Microbiology (2023)
-
Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR
Nature Biotechnology (2023)
-
CRISPR/Cas9-mediated gene knockout and interallelic gene conversion in human induced pluripotent stem cells using non-integrative bacteriophage-chimeric retrovirus-like particles
BMC Biology (2022)
-
Formation of synthetic RNA protein granules using engineered phage-coat-protein -RNA complexes
Nature Communications (2022)
-
Single-cell measurement of plasmid copy number and promoter activity
Nature Communications (2021)