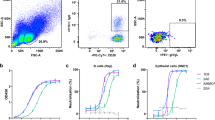
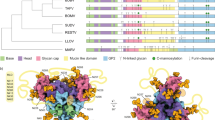
Abstract
Epstein-Barr virus (EBV) infection of B cells is associated with lymphoma and other human cancers. EBV infection is initiated by the binding of the viral envelope glycoprotein (gp350) to the cell surface receptor CR2. We determined the X-ray structure of the highly glycosylated gp350 and defined the CR2 binding site on gp350. Polyglycans shield all but one surface of the gp350 polypeptide, and we demonstrate that this glycan-free surface is the receptor-binding site. Deglycosylated gp350 bound CR2 similarly to the glycosylated form, suggesting that glycosylation is not important for receptor binding. Structure-guided mutagenesis of the glycan-free surface disrupted receptor binding as well as binding by a gp350 monoclonal antibody, a known inhibitor of virus-receptor interactions. These results provide structural information for developing drugs and vaccines to prevent infection by EBV and related viruses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Miller, R.F., Jones, E.L., Duddy, M.J. & Shahmanesh, M. Progressive intrathoracic lymphadenopathy: EBV associated non-Hodgkin's lymphoma. Sex. Transm. Infect. 78, 13–17 (2002).
Serraino, D. et al. Infection with Epstein-Barr virus and cancer: an epidemiological review. J. Biol. Regul. Homeost. Agents 19, 63–70 (2005).
Gandhi, M.K. Epstein-Barr virus-associated lymphomas. Expert Rev. Anti Infect. Ther. 4, 77–89 (2006).
Young, L.S. & Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4, 757–768 (2004).
Griffin, B.E. Epstein-Barr virus (EBV) and human disease: facts, opinions and problems. Mutat. Res. 462, 395–405 (2000).
Fingeroth, J.D. et al. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 81, 4510–4514 (1984).
Tanner, J., Weis, J., Fearon, D., Whang, Y. & Kieff, E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50, 203–213 (1987).
Ahearn, J.M., Hayward, S.D., Hickey, J.C. & Fearon, D.T. Epstein-Barr virus (EBV) infection of murine L cells expressing recombinant human EBV/C3d receptor. Proc. Natl. Acad. Sci. USA 85, 9307–9311 (1988).
Molesworth, S.J., Lake, C.M., Borza, C.M., Turk, S.M. & Hutt-Fletcher, L.M. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74, 6324–6332 (2000).
Mullen, M.M., Haan, K.M., Longnecker, R. & Jardetzky, T.S. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9, 375–385 (2002).
Weis, J.J., Tedder, T.F. & Fearon, D.T. Identification of a 145,000 Mr membrane protein as the C3d receptor (CR2) of human B lymphocytes. Proc. Natl. Acad. Sci. USA 81, 881–885 (1984).
Szakonyi, G. et al. Structure of complement receptor 2 in complex with its C3d ligand. Science 292, 1725–1728 (2001).
Sarrias, M.R. et al. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J. Immunol. 167, 1490–1499 (2001).
Guthridge, J.M. et al. Epitope mapping using the X-ray crystallographic structure of complement receptor type 2 (CR2/CD21): identification of a highly inhibitory monoclonal antibody that directly recognizes the CR2–C3d interface. J. Immunol. 167, 5758–5766 (2001).
Tanner, J., Whang, Y., Sample, J., Sears, A. & Kieff, E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 62, 4452–4464 (1988).
Martin, D.R., Yuryev, A., Kalli, K.R., Fearon, D.T. & Ahearn, J.M. Determination of the structural basis for selective binding of Epstein-Barr virus to human complement receptor type 2. J. Exp. Med. 174, 1299–1311 (1991).
Lowell, C.A. et al. Mapping of the Epstein-Barr virus and C3dg binding sites to a common domain on complement receptor type 2. J. Exp. Med. 170, 1931–1946 (1989).
Nemerow, G.R., Mullen, J.J., III, Dickson, P.W. & Cooper, N.R. Soluble recombinant CR2 (CD21) inhibits Epstein-Barr virus infection. J. Virol. 64, 1348–1352 (1990).
Hebell, T., Ahearn, J.M. & Fearon, D.T. Suppression of the immune response by a soluble complement receptor of B lymphocytes. Science 254, 102–105 (1991).
Jackman, W.T., Mann, K.A., Hoffmann, H.J. & Spaete, R.R. Expression of Epstein-Barr virus gp350 as a single chain glycoprotein for an EBV subunit vaccine. Vaccine 17, 660–668 (1999).
Emini, E.A., Schleif, W.A., Silberklang, M., Lehman, D. & Ellis, R.W. Vero cell-expressed Epstein-Barr virus (EBV) gp350/220 protects marmosets from EBV challenge. J. Med. Virol. 27, 120–123 (1989).
Finerty, S. et al. Protective immunization against Epstein-Barr virus-induced disease in cottontop tamarins using the virus envelope glycoprotein gp340 produced from a bovine papillomavirus expression vector. J. Gen. Virol. 73, 449–453 (1992).
Thorley-Lawson, D.A. & Geilinger, K. Monoclonal antibodies against the major glycoprotein (gp350/220) of Epstein-Barr virus neutralize infectivity. Proc. Natl. Acad. Sci. USA 77, 5307–5311 (1980).
Metzler, W.J. et al. Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28. Nat. Struct. Biol. 4, 527–531 (1997).
Hsu, T.A. et al. Differential N-glycan patterns of secreted and intracellular IgG produced in Trichoplusia ni cells. J. Biol. Chem. 272, 9062–9070 (1997).
Hannan, J.P. et al. Mutational analysis of the complement receptor type 2 (CR2/CD21)-C3d interaction reveals a putative charged SCR1 binding site for C3d. J. Mol. Biol. 346, 845–858 (2005).
Nagar, B., Jones, R.G., Diefenbach, R.J., Isenman, D.E. & Rini, J.M. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science 280, 1277–1281 (1998).
Prota, A.E., Sage, D.R., Stehle, T. & Fingeroth, J.D. The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding. Proc. Natl. Acad. Sci. USA 99, 10641–10646 (2002).
Nemerow, G.R., Houghten, R.A., Moore, M.D. & Cooper, N.R. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56, 369–377 (1989).
Urquiza, M., Lopez, R., Patino, H., Rosas, J.E. & Patarroyo, M.E. Identification of three gp350/220 regions involved in Epstein-Barr virus invasion of host cells. J. Biol. Chem. 280, 35598–35605 (2005).
Zhang, P.F. & Marcus-Sekura, C.J. Conformation-dependent recognition of baculovirus-expressed Epstein-Barr virus gp350 by a panel of monoclonal antibodies. J. Gen. Virol. 74, 2171–2179 (1993).
Pither, R.J., Nolan, L., Tarlton, J., Walford, J. & Morgan, A.J. Distribution of epitopes within the amino acid sequence of the Epstein-Barr virus major envelope glycoprotein, gp340, recognized by hyperimmune rabbit sera. J. Gen. Virol. 73, 1409–1415 (1992).
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
de la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–493 (1997).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Acknowledgements
The work was supported by US National Institutes of Health grants R01AI050096 to X.S.C. and R0-1 R01CA053615 to V.M.H. We thank staff scientists at the 19ID beamline at the Structural Biology Center in Argonne National Laboratory and at Advanced Light Source beamlines BL8.2.1 and BL 8.2.2 for assistance in data collection.
Author information
Authors and Affiliations
Contributions
G.S. purified the proteins, grew the crystals, collected data, solved the phases and built the initial model. M.G.K. improved the phases and refined the model. J.P.H. and K.A.Y. made the mutants and conducted the binding studies. R.Z.M. generated antibodies for cloning and expression of gp350. R.A. helped with the initial protein purification. V.M.H. participated in the experimental design and data analysis. X.S.C. supervised the entire project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Szakonyi, G., Klein, M., Hannan, J. et al. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol 13, 996–1001 (2006). https://doi.org/10.1038/nsmb1161
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1161
This article is cited by
-
Lymph node targeted multi-epitope subunit vaccine promotes effective immunity to EBV in HLA-expressing mice
Nature Communications (2023)
-
Preparation of monoclonal antibodies against Epstein-Barr virus glycoprotein 350
Virus Genes (2023)
-
A high-throughput neutralizing assay for antibodies and sera evaluation against Epstein-Barr virus
Virology Journal (2022)
-
Functional diversity: update of the posttranslational modification of Epstein–Barr virus coding proteins
Cellular and Molecular Life Sciences (2022)
-
The structural basis of herpesvirus entry
Nature Reviews Microbiology (2021)