Abstract
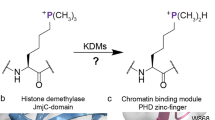
The reversible methylation of specific lysine residues in histone tails is crucial in epigenetic gene regulation. LSD1, the first known lysine-specific demethylase, selectively removes monomethyl and dimethyl, but not trimethyl modifications of Lys4 or Lys9 of histone-3. Here, we present the crystal structure of LSD1 at 2.9-Å resolution. LSD1 forms a highly asymmetric, closely packed domain structure from which a long helical 'tower' domain protrudes. The active site cavity is spacious enough to accommodate several residues of the histone tail substrate, but does not appear capable of recognizing the different methylation states of the substrate lysine. This supports the hypothesis that trimethylated lysine is chemically rather than sterically discriminated. We present a biochemical analysis of LSD1 mutants that identifies crucial residues in the active site cavity and shows the importance of the SWIRM and tower domains for catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Lee, M.G., Wynder, C., Cooch, N. & Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005).
Shi, Y.J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Wysocka, J., Milne, T.A. & Allis, C.D. Taking LSD1 to a new high. Cell 122, 654–658 (2005).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Kubicek, S. & Jenuwein, T. A crack in histone lysine methylation. Cell 119, 903–906 (2004).
Aravind, L. & Iyer, L.M. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 3, RESEARCH0039 (2002).
Forneris, F., Binda, C., Vanoni, M.A., Mattevi, A. & Battaglioli, E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 579, 2203–2207 (2005).
Forneris, F., Binda, C., Vanoni, M.A., Battaglioli, E. & Mattevi, A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280, 41360–41365 (2005).
Qian, C. et al. Structure and chromosomal DNA binding of the SWIRM domain. Nat. Struct. Mol. Biol. 12, 1078–1085 (2005).
Da, G. et al. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl. Acad. Sci. USA 103, 2057–2062 (2006).
Binda, C., Angelini, R., Federico, R., Ascenzi, P. & Mattevi, A. Structural bases for inhibitor binding and catalysis in polyamine oxidase. Biochemistry 40, 2766–2776 (2001).
Bannister, A.J. & Kouzarides, T. Reversing histone methylation. Nature 436, 1103–1106 (2005).
Fraaije, M.W. & Mattevi, A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 (2000).
Hoelz, A., Nairn, A.C. & Kuriyan, J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol. Cell 11, 1241–1251 (2003).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography, Acta Crystallogr. D Biol. Crystallogr, 50, 760–763, (1994).
Murakami, K.S., Masuda, S., Campbell, E.A., Muzzin, O. & Darst, S.A. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296, 1285–1290 (2002).
La Fortelle, E.D. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 276, 476–494 (1997).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Sanner, M.F., Olson, A.J. & Spehner, J.C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Jeanmougin, F., Thompson, J.D., Guoy, M., Higgins, D.G. & Gibson, T.J. Multiple sequence alignment with Clustal X. Trends Biochem. 23, 403–405 (1998).
Acknowledgements
We thank the Kazusa DNA Research Institute for the complementary DNA clone of human LSD1, C. Ralston for invaluable support during data collection at the Advanced Light Source, K. Heyman for technical support, A. Patke for discussions, S. Lawrie and S. Etherton for help with editing the manuscript and members of the Blobel laboratory for discussions and comments on the manuscript. N-terminal protein sequencing and peptide synthesis were performed by the Protein Center of the Rockefeller University, and multiangle light-scattering analysis was carried out by the Yale Proteomics Facility. A.H. was supported by a grant from the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Contributions
P.S. and A.H. designed and performed experiments; P.S., G.B. and A.H. analyzed and interpreted data; A.H. prepared the figures and the manuscript.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Sequence alignment of LSD1 homologs (PDF 219 kb)
Supplementary Fig. 2
Electron density maps (PDF 5025 kb)
Supplementary Fig. 3
The SWIRM domain of LSD1 (PDF 251 kb)
Supplementary Fig. 4
Trimethylated H3-K4 peptide acts as a competitive inhibitor of LSD1 activity (PDF 50 kb)
Supplementary Table 1
Biochemical analysis of LSD1 mutants (PDF 477 kb)
Supplementary Table 2
Crystallographic analysis (PDF 461 kb)
Rights and permissions
About this article
Cite this article
Stavropoulos, P., Blobel, G. & Hoelz, A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 13, 626–632 (2006). https://doi.org/10.1038/nsmb1113
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1113
This article is cited by
-
Epigenetic modifications in the development of bronchopulmonary dysplasia: a review
Pediatric Research (2024)
-
LSD1: an emerging face in altering the tumor microenvironment and enhancing immune checkpoint therapy
Journal of Biomedical Science (2023)
-
A developmental role for the chromatin-regulating CoREST complex in the cnidarian Nematostella vectensis
BMC Biology (2022)
-
Histone demethylase Lsd1 is required for the differentiation of neural cells in Nematostella vectensis
Nature Communications (2022)
-
1H NMR-based assay for lysine demethylase LSD1 and its application to inhibitor screening
Genome Instability & Disease (2021)