Abstract
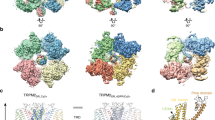
TRPML3 channels are mainly localized to endolysosomes and play a critical role in the endocytic pathway. Their dysfunction causes deafness and pigmentation defects in mice. TRPML3 activity is inhibited by low endolysosomal pH. Here we present cryo-electron microscopy (cryo-EM) structures of human TRPML3 in the closed, agonist-activated, and low-pH-inhibited states, with resolutions of 4.06, 3.62, and 4.65 Å, respectively. The agonist ML-SA1 lodges between S5 and S6 and opens an S6 gate. A polycystin-mucolipin domain (PMD) forms a luminal cap. S1 extends into this cap, forming a 'gating rod' that connects directly to a luminal pore loop, which undergoes dramatic conformational changes in response to low pH. S2 extends intracellularly and interacts with several intracellular regions to form a 'gating knob'. These unique structural features, combined with the results of electrophysiological studies, indicate a new mechanism by which luminal pH and other physiological modulators such as PIP2 regulate TRPML3 by changing S1 and S2 conformations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Di Fiore, P.P. & von Zastrow, M. Endocytosis, signaling, and beyond. Cold Spring Harb. Perspect. Biol. 6, a016865 (2014).
Xu, H., Martinoia, E. & Szabo, I. Organellar channels and transporters. Cell Calcium 58, 1–10 (2015).
Grimm, C., Butz, E., Chen, C.-C., Wahl-Schott, C. & Biel, M. From mucolipidosis type IV to Ebola: TRPML and two-pore channels at the crossroads of endo-lysosomal trafficking and disease. Cell Calcium 67, 148–155 (2017).
Venkatachalam, K., Wong, C.O. & Zhu, M.X. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium 58, 48–56 (2015).
Xu, H. & Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015).
Grimm, C., Barthmes, M. & Wahl-Schott, C. TRPML3. Handb. Exp. Pharmacol. 222, 659–674 (2014).
Noben-Trauth, K. The TRPML3 channel: from gene to function. Adv. Exp. Med. Biol. 704, 229–237 (2011).
Grimm, C., Hassan, S., Wahl-Schott, C. & Biel, M. Role of TRPML and two-pore channels in endolysosomal cation homeostasis. J. Pharmacol. Exp. Ther. 342, 236–244 (2012).
Li, M. et al. Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat. Struct. Mol. Biol. 24, 205–213 (2017).
Grieben, M. et al. Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2). Nat. Struct. Mol. Biol. 24, 114–122 (2017).
Shen, P.S. et al. The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell 167, 763–773 (2016).
Wilkes, M. et al. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat. Struct. Mol. Biol. 24, 123–130 (2017).
Kiselyov, K. et al. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 280, 43218–43223 (2005).
Dong, X.P. et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996 (2008).
Dong, X.P. et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284, 32040–32052 (2009).
Dong, X.P. et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 1, 38 (2010).
Dong, X.P., Wang, X. & Xu, H. TRP channels of intracellular membranes. J. Neurochem. 113, 313–328 (2010).
Li, X. et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016).
Wang, W. et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. USA 112, E1373–E1381 (2015).
Cheng, X. et al. The intracellular Ca2+ channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nat. Med. 20, 1187–1192 (2014).
Miedel, M.T. et al. Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 205, 1477–1490 (2008).
Samie, M. et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell 26, 511–524 (2013).
Zhang, X. et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 7, 12109 (2016).
LaPlante, J.M. et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89, 339–348 (2006).
Venkatachalam, K. et al. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell 135, 838–851 (2008).
Vergarajauregui, S., Connelly, P.S., Daniels, M.P. & Puertollano, R. Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 17, 2723–2737 (2008).
Garrity, A.G. et al. The endoplasmic reticulum, not the pH gradient, drives calcium refilling of lysosomes. eLife 5, e15887 (2016).
Sahoo, N. et al. Gastric acid secretion from parietal cells is mediated by a Ca2+ efflux channel in the tubulovesicle. Dev. Cell 41, 262–273 (2017).
Sun, M. et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9, 2471–2478 (2000).
Bargal, R. et al. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26, 118–123 (2000).
Bassi, M.T. et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin, and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67, 1110–1120 (2000).
Weitz, R. & Kohn, G. Clinical spectrum of mucolipidosis type IV. Pediatrics 81, 602–603 (1988).
Bach, G. Mucolipidosis type IV. Mol. Genet. Metab. 73, 197–203 (2001).
Wakabayashi, K., Gustafson, A.M., Sidransky, E. & Goldin, E. Mucolipidosis type IV: an update. Mol. Genet. Metab. 104, 206–213 (2011).
Di Palma, F. et al. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA 99, 14994–14999 (2002).
Grimm, C. et al. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc. Natl. Acad. Sci. USA 104, 19583–19588 (2007).
Kim, H.J. et al. Gain-of-function mutation in TRPML3 causes the mouse varitint-waddler phenotype. J. Biol. Chem. 282, 36138–36142 (2007).
Nagata, K. et al. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc. Natl. Acad. Sci. USA 105, 353–358 (2008).
Xu, H., Delling, M., Li, L., Dong, X. & Clapham, D.E. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. USA 104, 18321–18326 (2007).
Stauber, T. & Jentsch, T.J. Chloride in vesicular trafficking and function. Annu. Rev. Physiol. 75, 453–477 (2013).
Kiselyov, K.K., Ahuja, M., Rybalchenko, V., Patel, S. & Muallem, S. The intracellular Ca2+ channels of membrane traffic. Channels (Austin) 6, 344–351 (2012).
Patel, S. & Cai, X. Evolution of acidic Ca2+ stores and their resident Ca2+-permeable channels. Cell Calcium 57, 222–230 (2015).
Appelqvist, H., Wäster, P., Kågedal, K. & Öllinger, K. The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell Biol. 5, 214–226 (2013).
Di Paolo, G. & De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 (2006).
Zhang, X., Li, X. & Xu, H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl. Acad. Sci. USA 109, 11384–11389 (2012).
Raychowdhury, M.K. et al. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum. Mol. Genet. 13, 617–627 (2004).
Cantiello, H.F. et al. Cation channel activity of mucolipin-1: the effect of calcium. Pflugers Arch. 451, 304–312 (2005).
Kim, H.J. et al. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 27, 1197–1205 (2008).
Grimm, C., Jörs, S., Guo, Z., Obukhov, A.G. & Heller, S. Constitutive activity of TRPML2 and TRPML3 channels versus activation by low extracellular sodium and small molecules. J. Biol. Chem. 287, 22701–22708 (2012).
Wang, X. et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012).
Miao, Y., Li, G., Zhang, X., Xu, H. & Abraham, S.N. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell 161, 1306–1319 (2015).
Grimm, C. et al. Small molecule activators of TRPML3. Chem. Biol. 17, 135–148 (2010).
Shen, D. et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3, 731 (2012).
Feng, X., Xiong, J., Lu, Y., Xia, X. & Zhu, M.X. Differential mechanisms of action of the mucolipin synthetic agonist, ML-SA1, on insect TRPML and mammalian TRPML1. Cell Calcium 56, 446–456 (2014).
Zhang, S., Li, N., Zeng, W., Gao, N. & Yang, M. Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism. Protein Cell http://dx.doi.org/10.1007/s13238-017-0476-5 (2017).
Kim, H.J., Yamaguchi, S., Li, Q., So, I. & Muallem, S. Properties of the TRPML3 channel pore and its stable expansion by the varitint-waddler-causing mutation. J. Biol. Chem. 285, 16513–16520 (2010).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Biffi, A. Gene therapy for lysosomal storage disorders: a good start. Hum. Mol. Genet. 25, R65–R75 (2016).
Zheng, S.Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Li, M. et al. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65 (2017).
Bharat, T.A., Russo, C.J., Löwe, J., Passmore, L.A. & Scheres, S.H. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753 (2015).
Kucukelbir, A., Sigworth, F.J. & Tagare, H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Smart, O.S., Neduvelil, J.G., Wang, X., Wallace, B.A. & Sansom, M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Acknowledgements
This work was supported by the National Basic Research Program of China (grant 2014CB910301 to J.Y.), the National Institutes of Health (grant R01GM085234 to J.Y.), the National Natural Science Foundation of China (grant 31370821 to J.Y.; grant 31570730 to X.L.), the National Key Research and Development Program (grants 2016YFA0501102 and 2016YFA0501902 to X.L.), the Top Talents Program of Yunnan Province (grant 2011HA012 to J.Y.), the High-level Overseas Talents of Yunnan Province (J.Y.), the China Youth 1000-Talent Program of the State Council of China (X.L.), Beijing Advanced Innovation Center for Structural Biology (X.L.), and the Tsinghua-Peking Joint Center for Life Sciences (X.L.).
Author information
Authors and Affiliations
Contributions
M.L. and J.Y. conceived and initiated the project. X.Z., M.L., D.S., Q.J., H.L., X.L., and J.Y. designed the experiments, analyzed the results, and wrote the manuscript. M.L. performed all molecular biology and biochemical experiments and built the atomic models. X.Z. and X.L. performed all cryo-EM experiments, including data acquisition and processing. D.S., Q.J., and H.L. performed electrophysiology experiments. All authors contributed to manuscript discussion, preparation, and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Amino acid sequence alignment of human TRPML subunits.
Secondary structural elements are marked according to the pH 7.4 ML-SA1-bound TRPML3 structure. Green and yellow highlight identical and similar amino acids, respectively. The ion selectivity filter is boxed in red. The luminal pore-loop is boxed in blue, with the luminal pore-loop aspartates marked by red triangles. Red and green circles mark amino acids involved in binding PI(4,5)P2 and PI(3,5)P2, respectively. H283 is indicated by a red star. Orange circles mark amino acids involved in binding ML-SA1. Yellow star marks A419.
Supplementary Figure 2 Single-particle cryo-EM analysis of apo-TRPML3 at pH 7.4.
(a) A representative motion-corrected micrograph. Typical particles are marked with orange boxes. (b) Fourier power spectrum of the micrograph shown in a. (c) Gallery of typical two-dimensional class averages. (d) Flow chart of cryo-EM data processing. (e) Euler angle distribution of all particles used in the final map reconstruction. Each orientation is represented by a cylinder, for which both the height and color (from blue to red) are proportional to the number of particles for that specific direction. (f) Local resolution of the cryo-density map. (g) The gold-standard FSC curve of the final reconstruction (black) and the FSC curve between the final reconstruction and the map calculated from the atom model (blue). (h) Model validation. Blue, model versus the summed map. Black, model versus half 1 map (called ‘work’, used for model refinement). Red, model versus half 2 map (called ‘free’, not used for model refinement).
Supplementary Figure 3 Single-particle cryo-EM analysis of ML-SA1-bound TRPML3 at pH 7.4.
(a) A representative motion-corrected micrograph. Typical particles are marked with orange boxes. (b) Fourier power spectrum of the micrograph shown in a. (c) Gallery of typical two-dimensional class averages. (d) Flow chart of cryo-EM data processing. (e) Euler angle distribution of all particles used in the final map reconstruction. Each orientation is represented by a cylinder, for which both the height and color (from blue to red) are proportional to the number of particles for that specific direction. (f) Local resolution of the cryo-density map. (g) The FSC curve of the final reconstruction (black) and the FSC curve between the final reconstruction and the map calculated from the atom model (blue). (h) Model validation. Blue, model versus the summed map. Black, model versus half 1 map (called ‘work’, used for model refinement). Red, model versus half 2 map (called ‘free’, not used for model refinement).
Supplementary Figure 4 Single-particle cryo-EM analysis of apo-TRPML3 at pH 4.8.
(a) A representative motion-corrected micrograph. Typical particles are marked with orange boxes. (b) Fourier power spectrum of the micrograph shown in a. (c) Gallery of typical two-dimensional class averages. (d) Flow chart of cryo-EM data processing. (e) Euler angle distribution of all particles used in the final map reconstruction. Each orientation is represented by a cylinder, for which both the height and color (from blue to red) are proportional to the number of particles for that specific direction. (f) Local resolution of the cryo-density map. (g) The FSC curve of the final reconstruction (black) and the FSC curve between the final reconstruction and the map calculated from the atom model (blue). (h) Model validation. Blue, model versus the summed map. Black, model versus half 1 map (called ‘work’, used for model refinement). Red, model versus half 2 map (called ‘free’, not used for model refinement).
Supplementary Figure 5 Representative cryo-EM density maps.
(a) Cryo-EM density maps and atomic models of selected regions of TRPML3 in the ML-SA1-bound pH 7.4 condition (left) or the apo pH 4.8 condition (right). The ML-SA1-bound pH 7.4 maps were low-pass filtered to 3.62 Å and amplified by a temperature factor of -180 Å2, and were contoured at 3.0σ. The apo pH 4.8 maps were low-pass filtered to 4.65 Å and amplified by a temperature factor of -244 Å2, and were contoured at 4.0σ. (b) Comparison of the cryo-EM density map in a crevice surrounded by S5, S6 and pore helix 1 in the apo pH 7.4 (left, filtered to 4.06 Å and contoured at 3.0σ) and ML-SA1-bound pH 7.4 (middle, filtered to 3.62 Å and contoured at 3.0σ) structures. The right panel shows the normalized different density map between the two structures (filtered to 6 Å and contoured at 15.0σ).
Supplementary Figure 6 Structure of the TRPML3 PMD compared with that of the TRPML1 PMD.
(a) Electrostatic-potential surface representation of the TRPML3 PMD, viewed from the luminal side of the membrane (left) or parallel to the membrane (right). (b) Superposition of TRPML3 and TRPML1 PMDs. (c). Superposition of the backbone α carbons of the two luminal pore-loops in stereo view. Same color representation as in b. The first amino acid of each luminal pore-loop is numbered 1. (d) Stereo view of the TRPML3 luminal pore-loop. (e) Stereo view of the TRPML1 luminal pore-loop.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 2571 kb)
Life Sciences Reporting Summary
Original electrophysiological data for Figures 2 and 5 (PDF 130 kb)
Supplementary Data Set 1
Source data for Figures 2 and 5. (XLSX 95 kb)
Conformational changes induced by the binding of ML-SA1
ML-SA1 binding causes many movements. For example, when viewed from the side, S5 and S6 move outward and the S4-S5 linker moves downward by 2 to 4 Å, the pore-loop moves downward by ∼2 Å, and S6 undergoes a 27 degree counterclockwise rotation. The zoom-in view from the bottom shows the movement of I498 (in space-filling form) between the closed state and open state upon ML-SA1 binding and unbinding. (MOV 23233 kb)
Conformational changes induced by pH changes
The movie shows a morph between the pH 7.4 apo structure and the pH 4.8 apo structure. The channel is viewed first from the side (parallel to the membrane) and then from top down (perpendicular to the membrane). (MOV 16106 kb)
Rights and permissions
About this article
Cite this article
Zhou, X., Li, M., Su, D. et al. Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nat Struct Mol Biol 24, 1146–1154 (2017). https://doi.org/10.1038/nsmb.3502
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3502
This article is cited by
-
StarMap: a user-friendly workflow for Rosetta-driven molecular structure refinement
Nature Protocols (2023)
-
Structural basis for Ca2+ activation of the heteromeric PKD1L3/PKD2L1 channel
Nature Communications (2021)
-
Mechanism of ligand activation of a eukaryotic cyclic nucleotide−gated channel
Nature Structural & Molecular Biology (2020)
-
Adaptive selection drives TRPP3 loss-of-function in an Ethiopian population
Scientific Reports (2020)
-
Release and uptake mechanisms of vesicular Ca2+ stores
Protein & Cell (2019)