Abstract
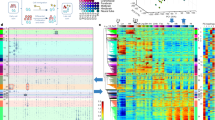
We determined genome-wide nucleosome occupancies in mouse embryonic stem cells and their neural progenitor and embryonic fibroblast counterparts to assess features associated with nucleosome positioning during lineage commitment. Cell-type- and protein-specific binding preferences of transcription factors to sites with either low (Myc, Klf4 and Zfx) or high (Nanog, Oct4 and Sox2) nucleosome occupancy as well as complex patterns for CTCF were identified. Nucleosome-depleted regions around transcription start and transcription termination sites were broad and more pronounced for active genes, with distinct patterns for promoters classified according to CpG content or histone methylation marks. Throughout the genome, nucleosome occupancy was correlated with certain histone methylation or acetylation modifications. In addition, the average nucleosome repeat length increased during differentiation by 5–7 base pairs, with local variations for specific regions. Our results reveal regulatory mechanisms of cell differentiation that involve nucleosome repositioning.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Sadeh, R. & Allis, C.D. Genome-wide “re”-modeling of nucleosome positions. Cell 147, 263–266 (2011).
Rando, O.J. & Winston, F. Chromatin and transcription in yeast. Genetics 190, 351–387 (2012).
Zhang, Z. & Pugh, B.F. High-resolution genome-wide mapping of the primary structure of chromatin. Cell 144, 175–186 (2011).
Cui, K. & Zhao, K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol. 833, 413–419 (2012).
Yuan, G.C. et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 (2005).
Ioshikhes, I.P., Albert, I., Zanton, S.J. & Pugh, B.F. Nucleosome positions predicted through comparative genomics. Nat. Genet. 38, 1210–1215 (2006).
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006).
Hu, G. et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 (2011).
Valouev, A. et al. Determinants of nucleosome organization in primary human cells. Nature 474, 516–520 (2011).
Ott, C.J. et al. Nucleosome occupancy reveals regulatory elements of the CFTR promoter. Nucleic Acids Res. 40, 625–637 (2012).
Schones, D.E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Zhang, L., Ma, H. & Pugh, B.F. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 21, 875–884 (2011).
Tirosh, I., Sigal, N. & Barkai, N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 11, R49 (2010).
Moshkin, Y.M. et al. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell Biol. 32, 675–688 (2012).
Li, Z., Schug, J., Tuteja, G., White, P. & Kaestner, K.H. The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 18, 742–746 (2011).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Martin, D. et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes. Nat. Struct. Mol. Biol. 18, 708–714 (2011).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366 (2009).
Schnetz, M.P. et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023 (2010).
Ho, L. et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 106, 5187–5191 (2009).
Zielke, N. et al. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123–127 (2011).
Teif, V.B. & Rippe, K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 37, 5641–5655 (2009).
Erdel, F., Krug, J., Langst, G. & Rippe, K. Targeting chromatin remodelers: signals and search mechanisms. Biochim. Biophys. Acta 1809, 497–508 (2011).
Cuddapah, S. et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 (2009).
Fu, Y., Sinha, M., Peterson, C.L. & Weng, Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4, e1000138 (2008).
Handoko, L. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 (2011).
Mavrich, T.N. et al. Nucleosome organization in the Drosophila genome. Nature 453, 358–362 (2008).
Valouev, A. et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18, 1051–1063 (2008).
Segal, E. & Widom, J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 19, 65–71 (2009).
Zhang, Y. et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16, 847–852 (2009).
Weiner, A., Hughes, A., Yassour, M., Rando, O.J. & Friedman, N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20, 90–100 (2010).
Fan, X. et al. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc. Natl. Acad. Sci. USA 107, 17945–17950 (2010).
Ku, M. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 (2008).
Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Lenhard, B., Sandelin, A. & Carninci, P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245 (2012).
Fan, Y. et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 (2005).
Cheng, C. & Gerstein, M. Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res. 40, 553–568 (2012).
Kanduri, M. et al. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell Biol. 22, 3339–3344 (2002).
Min, I.M. et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 25, 742–754 (2011).
Pham, C.D., Sims, H.I., Archer, T.K. & Schnitzler, G.R. Multiple distinct stimuli increase measured nucleosome occupancy around human promoters. PLoS ONE 6, e23490 (2011).
Zhao, J., Hyman, L. & Moore, C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999).
Schuster-Böckler, B. & Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 (2012).
Woodcock, C.L., Skoultchi, A.I. & Fan, Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006).
Fan, Y. et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell Biol. 23, 4559–4572 (2003).
Correll, S.J., Schubert, M.H. & Grigoryev, S.A. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 31, 2416–2426 (2012).
Mallm, J.P., Tschape, J.A., Hick, M., Filippov, M.A. & Muller, U.C. Generation of conditional null alleles for APP and APLP2. Genesis 48, 200–206 (2010).
Chung, H.R. et al. The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS ONE 5, e15754 (2010).
Cui, P. et al. A novel mechanism of epigenetic regulation: nucleosome-space occupancy. Biochem. Biophys. Res. Commun. 391, 884–889 (2010).
Allan, J., Fraser, R.M., Owen-Hughes, T. & Keszenman-Pereyra, D. Micrococcal nuclease does not substantially bias nucleosome mapping. J. Mol. Biol. 417, 152–164 (2012).
Xi, Y., Yao, J., Chen, R., Li, W. & He, X. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 21, 718–724 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Chereji, R.V., Tolkunov, D., Locke, G. & Morozov, A.V. Statistical mechanics of nucleosome ordering by chromatin-structure-induced two-body interactions. Phys. Rev. E 83, 050903 (2011).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Werner, T. Next generation sequencing in functional genomics. Brief. Bioinform. 11, 499–511 (2010).
Ward, J.H. Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 48, 236–244 (1963).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Caudron-Herger, M. et al. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus 2, 410–424 (2011).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Acknowledgements
We are grateful to A. Valouev and R. Chereji for help with the algorithms for calculations of NRL and average TSS patterns, respectively; to M. Gerstein for advice on data processing; to G. Längst, G. Wedemann and K. Fejes Tóth for discussions; and to the Deutsches Krebsforschungszentrum Sequencing Core Facility for conducting the sequencing. This work was funded within project EpiGenSys by the German Federal Ministry of Education and Research (BMBF) as a partner of the ERASysBio+ initiative in the EU FP7 ERA-NET Plus program through grant 0315712A to K.R. Computational resources and data storage were provided by grants from the BMBF (01IG07015G, Services@MediGRID) and the German Research Foundation (DFG INST 295/27-1). V.B.T. acknowledges the support from the Heidelberg Center for Modeling and Simulation in the Biosciences and a Deutsches Krebsforschungszentrum intramural grant, and Y.V. was supported by BMBF MedSys grant 0315409E to T.H.
Author information
Authors and Affiliations
Contributions
V.B.T. and K.R. designed the research. M.C.-H., J.-P.M. and C.M. performed experiments. V.B.T., Y.V., J.-P.M., T.H. and K.R. analyzed data. V.B.T., T.H. and K.R. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–4 and Supplementary Note (PDF 5299 kb)
Supplementary Table 5
Genes in cluster I of bivalent ESC promoters. (XLS 40 kb)
Supplementary Table 6
Differential promoter nucleosome occupancy and expression profiles of MEFs versus ESCs. (XLS 22311 kb)
Supplementary Table 7
Differential promoter nucleosome occupancy and expression profiles of NPCs versus ESCs. (XLS 24253 kb)
Rights and permissions
About this article
Cite this article
Teif, V., Vainshtein, Y., Caudron-Herger, M. et al. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19, 1185–1192 (2012). https://doi.org/10.1038/nsmb.2419
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2419
This article is cited by
-
Circulating cell-free DNA fragmentation is a stepwise and conserved process linked to apoptosis
BMC Biology (2023)
-
A conserved ZFX/WNT3 axis modulates the growth and imatinib response of chronic myeloid leukemia stem/progenitor cells
Cellular & Molecular Biology Letters (2023)
-
Histone exchange sensors reveal variant specific dynamics in mouse embryonic stem cells
Nature Communications (2023)
-
B1 SINE-binding ZFP266 impedes mouse iPSC generation through suppression of chromatin opening mediated by reprogramming factors
Nature Communications (2023)
-
DNA sequence-dependent formation of heterochromatin nanodomains
Nature Communications (2022)