Abstract
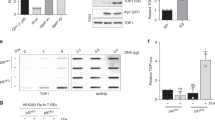
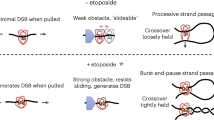
The topoisomerase II (topo II) DNA incision-and-ligation cycle can be poisoned (for example following treatment with cancer chemotherapeutics) to generate cytotoxic DNA double-strand breaks (DSBs) with topo II covalently conjugated to DNA. Tyrosyl-DNA phosphodiesterase 2 (Tdp2) protects genomic integrity by reversing 5′-phosphotyrosyl–linked topo II–DNA adducts. Here, X-ray structures of mouse Tdp2–DNA complexes reveal that Tdp2 β–2-helix–β DNA damage–binding 'grasp', helical 'cap' and DNA lesion–binding elements fuse to form an elongated protein-DNA conjugate substrate-interaction groove. The Tdp2 DNA-binding surface is highly tailored for engagement of 5′-adducted single-stranded DNA ends and restricts nonspecific endonucleolytic or exonucleolytic processing. Structural, mutational and functional analyses support a single–metal ion catalytic mechanism for the exonuclease-endonuclease-phosphatase (EEP) nuclease superfamily and establish a molecular framework for targeted small-molecule blockade of Tdp2-mediated resistance to anticancer topoisomerase drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Deweese, J.E. & Osheroff, N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 37, 738–748 (2009).
Wilstermann, A.M. & Osheroff, N. Base excision repair intermediates as topoisomerase II poisons. J. Biol. Chem. 276, 46290–46296 (2001).
Bandele, O.J. & Osheroff, N. (-)-Epigallocatechin gallate, a major constituent of green tea, poisons human type II topoisomerases. Chem. Res. Toxicol. 21, 936–943 (2008).
Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 (2009).
Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 (2009).
Hande, K.R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 34, 1514–1521 (1998).
Baldwin, E.L. & Osheroff, N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anticancer Agents 5, 363–372 (2005).
Cortes Ledesma, F., El Khamisy, S.F., Zuma, M.C., Osborn, K. & Caldecott, K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678 (2009).
Bahmed, K., Nitiss, K.C. & Nitiss, J.L. UnTTrapping the ends: a new player in overcoming protein linked DNA damage. Cell Res. 20, 122–123 (2010).
Mao, Y., Desai, S.D., Ting, C.Y., Hwang, J. & Liu, L.F. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 276, 40652–40658 (2001).
Fan, J.R. et al. Cellular processing pathways contribute to the activation of etoposide-induced DNA damage responses. DNA Repair (Amst.) 7, 452–463 (2008).
Do, P.M. et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 26, 830–845 (2012).
Zeng, Z., Cortes-Ledesma, F., El Khamisy, S.F. & Caldecott, K.W. TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem. 286, 403–409 (2011).
Wang, H. et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 29, 2566–2576 (2010).
Mol, C.D., Izumi, T., Mitra, S. & Tainer, J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 403, 451–456 (2000).
Pype, S. et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 275, 18586–18593 (2000).
Várady, G., Sarkadi, B. & Fatyol, K. TTRAP is a novel component of the non-canonical TRAF6–TAK1 TGF-β signaling pathway. PLoS ONE 6, e25548 (2011).
Garces, F., Pearl, L.H. & Oliver, A.W. The structural basis for substrate recognition by Mammalian polynucleotide kinase 3′ phosphatase. Mol. Cell 44, 385–396 (2011).
Schellenberg, M.J. & Williams, R.S. DNA end processing by polynucleotide kinase/phosphatase. Proc. Natl. Acad. Sci. USA 108, 20855–20856 (2011).
Bernstein, N.K. et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 17, 657–670 (2005).
Coquelle, N., Havali-Shahriari, Z., Bernstein, N., Green, R. & Glover, J.N. Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates. Proc. Natl. Acad. Sci. USA 108, 21022–21027 (2011).
Tumbale, P. et al. Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease. Nat. Struct. Mol. Biol. 18, 1189–1195 (2011).
Tsutakawa, S.E. et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145, 198–211 (2011).
Wu, C.C. et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 333, 459–462 (2011).
Schmidt, B.H., Burgin, A.B., Deweese, J.E., Osheroff, N. & Berger, J.M. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–644 (2010).
Hartsuiker, E., Neale, M.J. & Carr, A.M. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 (2009).
Nakamura, K. et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6, e1000828 (2010).
Neale, M.J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 (2005).
Williams, R.S., Williams, J.S. & Tainer, J.A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 (2007).
Davies, D.R., Interthal, H., Champoux, J.J. & Hol, W.G. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure 10, 237–248 (2002).
El-Khamisy, S.F. et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005).
Takashima, H. et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272 (2002).
Zeng, Z. et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 40, 8371–8380 (2012).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Stols, L. et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 (2002).
Otwinowski, Z. & Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. in Methods in Enzymology Vol. 276 (eds. Carter, C.W. Jr. & Sweets, R.M.) 307–326 Academic Press, 1997.
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Acknowledgements
This research was supported by the intramural research program of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS) grant 1Z01ES102765-02 (R.S.W.) and by NIH grant CA 084442 (D.A.R.). We thank T. Kunkel and members of the Williams lab for discussions and critical reading of the manuscript, L. Pedersen of the NIEHS Collaborative crystallography group, the Advanced Light Source beamline 12.3.1 (SIBYLS) staff and the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) staff for assistance with SAXS and crystallographic data collection and J. Williams of the NIEHS Protein Microcharacterization Core Facility for mass spectrometry analysis.
Author information
Authors and Affiliations
Contributions
M.J.S., C.D.A., P.D.R. and S.A. characterized the Tdp2 protein and enzymatic activity. C.D.A. and M.J.S. performed mutagenesis and analyzed mutant Tdp2 proteins. M.J.S. crystallized Tdp2 and Tdp2–DNA complexes. M.J.S. and R.S.W. collected SAXS and X-ray diffraction data and solved the X-ray crystal structures. M.J.S. and D.A.R. designed experiments, analyzed results and helped prepare the manuscript. R.S.W. conceived of and managed the study, designed experiments, analyzed results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4 (PDF 7575 kb)
Rights and permissions
About this article
Cite this article
Schellenberg, M., Appel, C., Adhikari, S. et al. Mechanism of repair of 5′-topoisomerase II–DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol 19, 1363–1371 (2012). https://doi.org/10.1038/nsmb.2418
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2418
This article is cited by
-
Inactivating TDP2 missense mutation in siblings with congenital abnormalities reminiscent of fanconi anemia
Human Genetics (2023)
-
Molecular mechanisms of topoisomerase 2 DNA–protein crosslink resolution
Cellular and Molecular Life Sciences (2020)
-
Topoisomerase-mediated chromosomal break repair: an emerging player in many games
Nature Reviews Cancer (2015)
-
Capturing snapshots of APE1 processing DNA damage
Nature Structural & Molecular Biology (2015)
-
TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function
Nature Genetics (2014)