Abstract
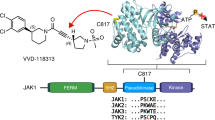
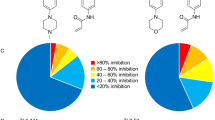
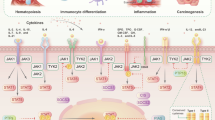
The protein tyrosine kinase JAK2 mediates signaling through numerous cytokine receptors. JAK2 possesses a pseudokinase domain (JH2) and a tyrosine kinase domain (JH1). Through unknown mechanisms, JH2 regulates the catalytic activity of JH1, and hyperactivating mutations in the JH2 region of human JAK2 cause myeloproliferative neoplasms (MPNs). We showed previously that JAK2 JH2 is, in fact, catalytically active. Here we present crystal structures of human JAK2 JH2, including both wild type and the most prevalent MPN mutant, V617F. The structures reveal that JH2 adopts the fold of a prototypical protein kinase but binds Mg-ATP noncanonically. The structural and biochemical data indicate that the V617F mutation rigidifies α-helix C in the N lobe of JH2, facilitating trans-phosphorylation of JH1. The crystal structures of JH2 afford new opportunities for the design of novel JAK2 therapeutics targeting MPNs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ghoreschi, K., Laurence, A. & O'Shea, J.J. Janus kinases in immune cell signaling. Immunol. Rev. 228, 273–287 (2009).
Levy, D.E. & Darnell, J.E. Jr. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 (2002).
Lucet, I.S. et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood 107, 176–183 (2006).
Boggon, T.J., Li, Y., Manley, P.W. & Eck, M.J. Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood 106, 996–1002 (2005).
Williams, N.K. et al. Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J. Mol. Biol. 387, 219–232 (2009).
Chrencik, J.E. et al. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J. Mol. Biol. 400, 413–433 (2010).
Vainchenker, W., Delhommeau, F., Constantinescu, S.N. & Bernard, O.A. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 118, 1723–1735 (2011).
Haan, C., Behrmann, I. & Haan, S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J. Cell. Mol. Med. 14, 504–527 (2010).
Kralovics, R. et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Baxter, E.J. et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005).
Levine, R.L. et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005).
Lipson, D. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 18, 382–384 (2012).
Russell, S.M. et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science 270, 797–800 (1995).
Saharinen, P. & Silvennoinen, O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 277, 47954–47963 (2002).
Saharinen, P., Takaluoma, K. & Silvennoinen, O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol. 20, 3387–3395 (2000).
Chen, M. et al. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol. Cell. Biol. 20, 947–956 (2000).
Lindauer, K., Loerting, T., Liedl, K.R. & Kroemer, R.T. Prediction of the structure of human Janus kinase 2 (JAK2) comprising the two carboxy-terminal domains reveals a mechanism for autoregulation. Protein Eng. 14, 27–37 (2001).
Ungureanu, D. et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol. 18, 971–976 (2011).
Zheng, J. et al. 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 49, 362–365 (1993).
Hubbard, S.R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16, 5572–5581 (1997).
Shi, F., Telesco, S.E., Liu, Y., Radhakrishnan, R. & Lemmon, M.A. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. USA 107, 7692–7697 (2010).
Jura, N., Shan, Y., Cao, X., Shaw, D.E. & Kuriyan, J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. USA 106, 21608–21613 (2009).
Dusa, A., Mouton, C., Pecquet, C., Herman, M. & Constantinescu, S.N. JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PLoS ONE 5, e11157 (2010).
Gnanasambandan, K., Magis, A. & Sayeski, P.P. The constitutive activation of Jak2–V617F is mediated by a π stacking mechanism involving phenylalanines 595 and 617. Biochemistry 49, 9972–9984 (2010).
Ni, Q., Shaffer, J. & Adams, J.A. Insights into nucleotide binding in protein kinase A using fluorescent adenosine derivatives. Protein Sci. 9, 1818–1827 (2000).
Argetsinger, L.S. et al. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol. Cell. Biol. 24, 4955–4967 (2004).
Ishida-Takahashi, R. et al. Phosphorylation of Jak2 on Ser(523) inhibits Jak2-dependent leptin receptor signaling. Mol. Cell. Biol. 26, 4063–4073 (2006).
Feener, E.P., Rosario, F., Dunn, S.L., Stancheva, Z. & Myers, M.G. Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol. Cell. Biol. 24, 4968–4978 (2004).
Mazurkiewicz-Munoz, A.M. et al. Phosphorylation of JAK2 at serine 523: a negative regulator of JAK2 that is stimulated by growth hormone and epidermal growth factor. Mol. Cell. Biol. 26, 4052–4062 (2006).
Zhao, L. et al. A JAK2 interdomain linker relays Epo receptor engagement signals to kinase activation. J. Biol. Chem. 284, 26988–26998 (2009).
Livnah, O. et al. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283, 987–990 (1999).
Tefferi, A. JAK inhibitors for myeloproliferative neoplasms: clarifying facts from myths. Blood 119, 2721–2730 (2012).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vagin, A.A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Painter, J. & Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
Hornak, V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006).
Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010).
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W. & Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Shaw, D.E. et al. Millisecond-scale molecular dynamics simulations on Anton. in ACAM/IEEE Conference on Supercomputing (ACM, 2009).
Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985).
Lippert, R.A. et al. A common, avoidable source of error in molecular dynamics integrators. J. Chem. Phys. 126, 046101 (2007).
Kräutler, V., Van Gunsteren, W.F. & Hunenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22, 501–508 (2001).
Fennell, C.J. & Gezelter, J.D. Is the Ewald summation still necessary? Pairwise alternatives to the accepted standard for long-range electrostatics. J. Chem. Phys. 124, 234104 (2006).
Acknowledgements
This work was supported in part by US National Institute of Allergy and Infectious Diseases grant R21 AI095808 (S.R.H.), the Medical Research Council of Academy of Finland (O.S.), the Sigrid Juselius Foundation (O.S.), the Finnish Cancer Foundation (O.S.), the Competitive Research Funding and Centre of Laboratory Medicine of the Tampere University Hospital (O.S.), the Maire Lisko Foundation (O.S.) and the Tampere Tuberculosis Foundation (O.S.). US National Cancer Institute training grant T32 CA009161 supported R.M.B. Financial support for beam line X25 at the National Synchrotron Light Source, Brookhaven National Laboratory, comes principally from the US Department of Energy, National Center for Research Resources (P41 RR012408) and National Institute of General Medical Sciences (P41 GM103473). We thank E. Koskenalho for technical assistance, V. DiGiacomo for peptide modeling, A. Héroux for X-ray data collection, W.T. Miller, M. Mohammadi and K. Gnanasambandan for critical reading of the manuscript and M. Eck (Dana-Farber Cancer Institute, Boston, Massachusetts, USA) for discussions and for providing coordinates before publication.
Author information
Authors and Affiliations
Contributions
R.M.B., crystallographic studies and manuscript preparation; D.U., in-cell biochemical studies and manuscript preparation; Y.S. and D.E.S., molecular dynamics simulations and manuscript preparation; O.S., project supervisor and manuscript preparation; S.R.H., project supervisor and principal manuscript author.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 1980 kb)
Rights and permissions
About this article
Cite this article
Bandaranayake, R., Ungureanu, D., Shan, Y. et al. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol 19, 754–759 (2012). https://doi.org/10.1038/nsmb.2348
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2348
This article is cited by
-
Tyrosine phosphorylation of CARM1 promotes its enzymatic activity and alters its target specificity
Nature Communications (2024)
-
Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer
Signal Transduction and Targeted Therapy (2023)
-
Preclinical studies of Flonoltinib Maleate, a novel JAK2/FLT3 inhibitor, in treatment of JAK2V617F-induced myeloproliferative neoplasms
Blood Cancer Journal (2022)
-
Role of water in the determination of protonation states of titratable residues
Journal of Molecular Modeling (2021)
-
MPL overexpression induces a high level of mutant-CALR/MPL complex: a novel mechanism of ruxolitinib resistance in myeloproliferative neoplasms with CALR mutations
International Journal of Hematology (2021)