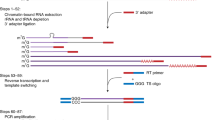
Abstract
It has recently been shown that RNA 3′-end formation plays a more widespread role in controlling gene expression than previously thought. To examine the impact of regulated 3′-end formation genome-wide, we applied direct RNA sequencing to A. thaliana. Here we show the authentic transcriptome in unprecedented detail and describe the effects of 3′-end formation on genome organization. We reveal extreme heterogeneity in RNA 3′ ends, discover previously unrecognized noncoding RNAs and propose widespread reannotation of the genome. We explain the origin of most poly(A)+ antisense RNAs and identify cis elements that control 3′-end formation in different registers. These findings are essential to understanding what the genome actually encodes, how it is organized and how regulated 3′-end formation affects these processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Di Giammartino, D.C., Nishida, K. & Manley, J.L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011).
Proudfoot, N.J. Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782 (2011).
Hornyik, C., Terzi, L.C. & Simpson, G.G. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18, 203–213 (2010).
Greger, I.H. & Proudfoot, N.J. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17, 4771–4779 (1998).
Gullerova, M., Moazed, D. & Proudfoot, N.J. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 25, 556–568 (2011).
Kuehner, J.N., Pearson, E.L. & Moore, C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12, 283–294 (2011).
Meyers, B.C. et al. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 22, 1006–1011 (2004).
Yamada, K. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003).
Stolc, V. et al. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc. Natl. Acad. Sci. USA 102, 4453–4458 (2005).
Wu, X. et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108, 12533–12538 (2011).
Gilboa, E., Mitra, S.W., Goff, S. & Baltimore, D. A detailed model of reverse transcription and tests of crucial aspects. Cell 18, 93–100 (1979).
Spiegelman, S. et al. DNA-directed DNA polymerase activity in oncogenic RNA viruses. Nature 227, 1029–1031 (1970).
Nam, D.K. et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. USA 99, 6152–6156 (2002).
Houseley, J. & Tollervey, D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE 5, e12271 (2010).
Perocchi, F., Xu, Z., Clauder-Munster, S. & Steinmetz, L.M. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 35, e128 (2007).
Jan, C.H., Friedman, R.C., Ruby, J.G. & Bartel, D.P. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97–101 (2011).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
Ozsolak, F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010).
Jin, Y. & Bian, T. Nontemplated nucleotide addition prior to polyadenylation: a comparison of Arabidopsis cDNA and genomic sequences. RNA 10, 1695–1697 (2004).
Loke, J.C. et al. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138, 1457–1468 (2005).
Mangone, M. et al. The landscape of C. elegans 3′UTRs. Science 329, 432–435 (2010).
Yepiskoposyan, H., Aeschimann, F., Nilsson, D., Okoniewski, M. & Muhlemann, O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17, 2108–2118 (2011).
Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell 136, 763–776 (2009).
Chekanova, J.A. et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353 (2007).
Brown, J.W., Echeverria, M. & Qu, L.H. Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 8, 42–49 (2003).
Wu, J.Q. et al. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 9, R3 (2008).
van Bakel, H., Nislow, C., Blencowe, B.J. & Hughes, T.R. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8, e1000371 (2010).
Okamura, K., Balla, S., Martin, R., Liu, N. & Lai, E.C. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 15, 581–590 (2008).
Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R. & Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291 (2005).
Katiyar-Agarwal, S. et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103, 18002–18007 (2006).
Kaufmann, I., Martin, G., Friedlein, A., Langen, H. & Keller, W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626 (2004).
Rothnie, H.M., Reid, J. & Hohn, T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 13, 2200–2210 (1994).
Sanfaçon, H., Brodmann, P. & Hohn, T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 5, 141–149 (1991).
Mayr, C. & Bartel, D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).
Huntzinger, E. & Izaurralde, E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110 (2011).
Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Prescott, E.M. & Proudfoot, N.J. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA 99, 8796–8801 (2002).
Henz, S.R. et al. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 144, 1247–1255 (2007).
Jen, C.H., Michalopoulos, I., Westhead, D.R. & Meyer, P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 6, R51 (2005).
Mapendano, C.K., Lykke-Andersen, S., Kjems, J., Bertrand, E. & Jensen, T.H. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol. Cell 40, 410–422 (2010).
Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011).
Ozsolak, F. et al. Direct RNA sequencing. Nature 461, 814–818 (2009).
Nicol, J.W., Helt, G.A., Blanchard, S.G. Jr., Raja, A. & Loraine, A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731 (2009).
Yang, J.H. et al. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 34, 5112–5123 (2006).
Chen, H.M. & Wu, S.H. Mining small RNA sequencing data: a new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 37, e69 (2009).
Kim, S.H. et al. Plant U13 orthologues and orphan snoRNAs identified by RNomics of RNA from Arabidopsis nucleoli. Nucleic Acids Res. 38, 3054–3067 (2010).
Barbezier, N. et al. Processing of a dicistronic tRNA-snoRNA precursor: combined analysis in vitro and in vivo reveals alternate pathways and coupling to assembly of snoRNP. Plant Physiol. 150, 1598–1610 (2009).
Acknowledgements
We thank T. Walsh for computing support, E. Bayne and R. Lyons for comments on the manuscript, Trivalent Editing and P. Smith for proofreading and the Biotechnology and Biological Sciences Research Council (BB/H002286/1) (A.S., C.D., C.C., G.J.B. and G.G.S.), the Scottish Government (C.H. and G.G.S.) and the US National Institutes of Health (HG005230 and HG005279) (F.O. and P.M.M.) for funding.
Author information
Authors and Affiliations
Contributions
G.G.S. and G.J.B. conceived and supervised the research. C.H. generated RNA samples. F.O. performed DRS. A.S. and C.C. analyzed the DRS data. C.D. and V.Z. did the molecular analyses of RNA. G.G.S. wrote the paper, and all authors read and commented on it.
Corresponding authors
Ethics declarations
Competing interests
F.O. and P.M.M. are employees of Helicos Biosciences Corporation.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 911 kb)
Supplementary Tables 1–12
Supplementary Tables 1–12 (XLS 7180 kb)
Rights and permissions
About this article
Cite this article
Sherstnev, A., Duc, C., Cole, C. et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19, 845–852 (2012). https://doi.org/10.1038/nsmb.2345
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2345
This article is cited by
-
Full-length transcriptome reconstruction reveals genetic differences in hybrids of Oryza sativa and Oryza punctata with different ploidy and genome compositions
BMC Plant Biology (2022)
-
Construction of drought stress regulation networks in potato based on SMRT and RNA sequencing data
BMC Plant Biology (2022)
-
In vivo nuclear RNA structurome reveals RNA-structure regulation of mRNA processing in plants
Genome Biology (2021)
-
Transcript isoform sequencing reveals widespread promoter-proximal transcriptional termination in Arabidopsis
Nature Communications (2020)
-
Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton
Nature Communications (2019)