Abstract
Human metapneumovirus and respiratory syncytial virus cause lower respiratory tract infections. The virus fusion (F) glycoprotein promotes membrane fusion by refolding from a metastable pre-fusion to a stable post-fusion conformation. F is also a major target of the neutralizing antibody response. Here we show that a potently neutralizing anti–human metapneumovirus antibody (DS7) binds a structurally invariant domain of F, revealing a new epitope that could be targeted in vaccine development.
Similar content being viewed by others
Main
Human metapneumovirus (HMPV) and respiratory syncytial virus (RSV)1,2,3 define the pneumovirus subfamily of the Paramyxoviridae and are major respiratory pathogens, causing substantial morbidity in infants and the elderly4,5. Vaccine development for HMPV and RSV has been challenging; there is currently no licensed vaccine for either virus. The paramyxovirus F protein is a class I viral fusion protein and a major target of the neutralizing antibody response6. F initially folds to a metastable, pre-fusion conformation6,7 that, upon activation, undergoes large-scale refolding6,7,8 that is coupled to membrane fusion. Although major antigenic sites in HMPV and RSV F have been identified, our understanding of F neutralizing epitopes remains incomplete9,10,11. The anti-HMPV F DS7 Fab was identified in a human antibody phage display library12,13, shows subnanomolar affinity and is highly protective in vivo9. We set out to understand how this neutralizing antibody interacts with HMPV F.
Previous EM studies have documented two conformations of the parainfluenzavirus 5 (PIV5), human PIV3 and Newcastle disease virus (NDV) F glycoproteins that match crystal structures of pre-fusion and post-fusion F conformations7,8,14,15. The pre-fusion HMPV F conformation was stabilized with a GCN4-derived trimerization tag, purified and visualized by negative stain EM (Fig. 1). Two trimeric forms of the intact F that are consistent with pre- and post-fusion states were observed14,15.
(a) The PIV5 F (pre-fusion) structure is shown in two views, rotated 90° from each other. One orientation shows the pre-fusion HRB stalk, and the second is oriented along the trimer axis and shows only the head region. The RSV F (post-fusion) trimer is shown, oriented perpendicularly to the trimer axis. (b) EM images of HMPV F that appear consistent with the pre-fusion (left) and post-fusion (right) conformational states. (c,d) EM images of DS7 Fab bound to HMPV F in putative pre-fusion (c) and post-fusion (d) conformations.
Purified DS7 Fab formed stable complexes with the HMPV F (Supplementary Fig. 1), and EM analysis revealed binding to the F head region (Fig. 1c,d). Three Fab molecules bound to the F trimer, which is particularly visible for molecules oriented along the F trimer axis that appear to represent pre-fusion HMPV F (Fig. 1c). Fab binding to the putative post-fusion conformation of HMPV F was also observed (Fig. 1d) where the F orientation creates an asymmetric 'T' shape.
DS7–F complexes prepared for crystallization showed spontaneous proteolysis, yielding an N-terminal, ~45-kDa fragment that crystallized with DS7 (Supplementary Figs. 1 and 2). The molecular weight of the complex (~110 kDa, Supplementary Fig. 1d) indicated that proteolysis led to dissociation of the trimer to a complex of one Fab bound to one F fragment. The structure was solved with heavy atom data and a Fab molecular-replacement solution (Supplementary Tables 1–3). The final model consists of HMPV F DI, DII and DIII domains bound to DS7 (refinement statistics are in Supplementary Table 1). DS7 and HMPV F DI–DII domains were completely modeled, whereas the F DIII lacked electron density for residues 95–113 and 165–177, corresponding to the fusion peptide and a connecting loop, respectively.
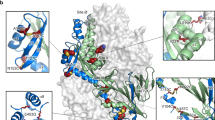
Consistent with the EM data, DS7 binds F residues located in the DI and DII head domains (Fig. 2). The interface buries ~1,600 Å2 of surface area involving 22 DS7 and 27 F residues. The DS7 heavy chain interacts exclusively with DI, contributing two-thirds of the total buried surface area. The heavy chain CDR3 inserts into a DI hydrophobic pocket formed by residues Leu24, Ile31, Pro282 and Trp284 (Fig. 2c). The DS7 light chain spans the DI–DII interface, with CDR1 engaging residues that form the first two strands of DI and three residues in DII.
(a,b) The DS7 Fab engages a single F fragment consisting of DI (yellow), DII (red) and DIII (magenta) domains. The DS7 heavy chain (light blue) binds a hydrophobic pocket in DI. The DS7 light chain (pale green) buries residues in both DI and a loop in DII. The DS7 heavy and light chain variable domains are indicated as VH and VL, respectively. The DS7 heavy and light chain constant domains are indicated as CH1 and CL, respectively. (b) The complex is rotated 90° from the orientation depicted in a. (c) Residues in the DS7 binding site interactions are shown in stick format, with carbon atoms colored by domain as in a. CDR3H inserts into a hydrophobic pocket on the broad side of DI that is formed by Leu24, Ile31, Pro282 and Trp284 and is flanked by charged residues Lys20, Glu33, Lys312, Arg348 and Glu349. The light chain buries residues at the DI N terminus and three residues in DII.
DS7 neutralizes representative strains of all HMPV genotypes (A1, A2, B1 and B2). Analysis of HMPV F sequence variability spanning a 20-year period reveals a mean of 96% amino acid identity across the four major subgroups16. F residues at the DS7 complex interface are highly conserved, with 25 of 27 identical amino acids in F proteins of A and B subgroups. The two F residues that vary (Gln312 or Lys312 and Lys348 or Arg348) flank the CDR3H binding pocket and appear to be compatible with binding (Fig. 2c).
The DS7 epitope does not involve residues within known HMPV or RSV F major antigenic sites (Fig. 3a). HMPV F sites fall into six groups, five of which have been mapped by selecting mAb-resistant virus mutants10,11,17, whereas six major antigenic sites (sites I –VI) have been identified for RSV F10,11,18. These major antigenic sites are located predominantly in DII and DIII, and they surround the DS7 epitope (Fig. 3a). The central region of DI that defines the majority of the DS7 epitope is markedly free of escape mutants, although it is a potent neutralizing site that should be exposed and conserved in structure in the pre- and post-fusion states.
(a) Major antigenic sites in HMPV and RSV F are mapped onto the HMPV F structure, with the DS7 buried surface area shown as a transparent yellow surface. The view is from the perspective of the DS7 Fab, rotated ~90° from that shown in Figure 2a. Escape-mutant sites are shown as side chains with dark blue (HMPV F) or magenta (RSV F) spheres. The individual HMPV residues are labeled, along with the HMPV antigenic sites II–VI and the RSV antigenic sites I–VI. The RSV antigenic sites correspond to the following HMPV residues (respective RSV sites in parentheses): Ag Site I: 357 (RSV: 389); Ag Site II: 232, 238, 242 and 245 (RSV: 262, 268, 272 and 275), Ag Sites IV–VI: 397, 400, 401, 404 and 415 (RSV: 429, 432, 433, 436 and 447). HMPV Site III includes residues 178 and 179. (b) The DS7 epitope surface is shown colored by atom type (yellow, carbon; blue, nitrogen and red, oxygen), with underlying residues shown in stick format. Asterisks mark residues conserved in HMPV and RSV F. A dotted line circumscribes a predominantly hydrophobic pocket that binds DS7 CDR3H. (c) The RSV F surface corresponding to the DS7 epitope is shown as in panel b. The dotted line highlights the partially conserved hydrophobic pocket with Trp314 at its base.
A comparison of HMPV and RSV F surfaces shows that crucial features of the DS7 epitope are also present in RSV, suggesting that this site could be a target for RSV vaccine development. Seven out of 28 amino acids are conserved in the two proteins, including the residues Pro282, Cys283, Trp284 and Asn351 (Fig. 3b,c and Supplementary Table 4). These residues, along with Ile31 in HMPV and Val40 in RSV, line a hydrophobic pocket, which in HMPV F engages the DS7 CDR3H and could correspond to a 'hot spot' for mAb binding to both proteins (Fig. 3b,c). In RSV, the pocket is partially filled in by the substitution of Leu24 with RSV Tyr33. Different amino acids surrounding the hydrophobic pocket would contribute to mAb specificity for RSV or HMPV F.
The refolding of paramyxovirus F proteins during membrane fusion and virus entry6,7 has a potentially major impact on neutralizing epitopes and thus antibody recognition. Neutralizing antibodies, such as DS7, palivizumab (or motavizumab) and 101F, recognize structural features that are mainly, if not completely, conserved in the pre- and post-fusion F conformations. Our DS7 complex structure highlights a neutralizing site that was not identified previously, and it may represent a low-frequency, subdominant antigenic site that is involved in the immune response to F. The position of the DS7 epitope, at the center of the structurally invariant DI domain and relatively distant from predicted subunit interfaces, suggests that DI domain- and scaffold-based vaccines might stimulate potent neutralizing responses to HMPV and RSV. Finally, we note that the HMPV F DIII retains a compact fold, similar to that of the pre-fusion PIV5 F DIII (Supplementary Figure 3), whereas the overall conformation of the three domains (DI-DII-DIII) better matches the post-fusion RSV F (Supplementary Discussion).
Accession code. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4DAG.
Accession codes
References
van den Hoogen, B.G. et al. Nat. Med. 7, 719–724 (2001).
Lamb, R.A. & Parks, G.D. in Fields Virology Vol. 1 (eds. Knipe, D.M. & Howley, P.M.) 1449–1496 (Lippincott Williams & Wilkins, 2007).
Collins, P.L. & Crowe, J.E. Jr. in Fields Virology Vol. 2 (eds. Knipe, D.M. & Howley, P.M.) 1601–1646 (Lippincott Williams & Wilkins, 2007).
Papenburg, J. & Boivin, G. Rev. Med. Virol. 20, 245–260 (2010).
Williams, J.V. et al. N. Engl. J. Med. 350, 443–450 (2004).
Lamb, R.A. & Jardetzky, T.S. Curr. Opin. Struct. Biol. 17, 427–436 (2007).
Yin, H.S., Wen, X., Paterson, R.G., Lamb, R.A. & Jardetzky, T.S. Nature 439, 38–44 (2006).
Yin, H.S., Paterson, R.G., Wen, X., Lamb, R.A. & Jardetzky, T.S. Proc. Natl. Acad. Sci. USA 102, 9288–9293 (2005).
Williams, J.V. et al. J. Virol. 81, 8315–8324 (2007).
Ulbrandt, N.D. et al. J. Gen. Virol. 89, 3113–3118 (2008).
Ulbrandt, N.D. et al. J. Virol. 80, 7799–7806 (2006).
Barbas, C.F. III et al. Proc. Natl. Acad. Sci. USA 89, 10164–10168 (1992).
Williamson, R.A. et al. Proc. Natl. Acad. Sci. USA 90, 4141–4145 (1993).
Connolly, S.A., Leser, G.P., Yin, H.S., Jardetzky, T.S. & Lamb, R.A. Proc. Natl. Acad. Sci. USA 103, 17903–17908 (2006).
Swanson, K. et al. Virology 402, 372–379 (2010).
Yang, C.F. et al. Virol. J. 6, 138 (2009).
Hamelin, M.E. et al. Antiviral Res. 88, 31–37 (2010).
Harbury, P.B., Kim, P.S. & Alber, T. Nature 371, 80–83 (1994).
Acknowledgements
We thank past and present members of the Jardetzky, Crowe, Williams and Lamb laboratories. This research was supported in part by US National Institutes of Health research grants to J.V.W. (AI-085062, AI-073697 and AI-072414), R.A.L. (AI-23173) and T.S.J. (GM-61050). R.A.L. is an investigator of the Howard Hughes Medical Institute. J.E.C. is a Burroughs Wellcome Fund Clinical Scientist in Translational Research.
Author information
Authors and Affiliations
Contributions
X.W., J.C.K., G.P.L., R.G.C., R.A.L., J.V.W., J.E.C. and T.S.J. designed experiments and reagents. X.W., J.C.K., G.P.L., R.G.C. and T.S.J. conducted experiments and contributed reagents. X.W., G.P.L. and T.S.J. wrote the manuscript and Supplementary information and prepared the figures. X.W., J.C.K., G.P.L., R.G.C., R.A.L., J.V.W., J.E.C. and T.S.J. analyzed data and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–4 and Supplementary Discussion (PDF 816 kb)
Rights and permissions
About this article
Cite this article
Wen, X., Krause, J., Leser, G. et al. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol 19, 461–463 (2012). https://doi.org/10.1038/nsmb.2250
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2250
This article is cited by
-
Cross-protective antibodies against common endemic respiratory viruses
Nature Communications (2023)
-
Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins
Nature Communications (2022)
-
Profiling of hMPV F-specific antibodies isolated from human memory B cells
Nature Communications (2022)
-
Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus
Nature Microbiology (2017)
-
Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein
Nature Communications (2017)