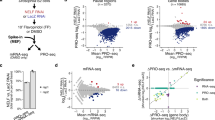
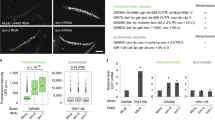
Abstract
Endogenous RNA interference (endo-RNAi) pathways use a variety of mechanisms to generate siRNA and to mediate gene silencing. In Caenorhabditis elegans, DCR-1 is essential for competing RNAi pathways—the ERI endo-RNAi pathway and the exogenous RNAi pathway—to function. Here, we demonstrate that DCR-1 forms exclusive complexes in each pathway and further define the ERI–DCR-1 complex. We show that the tandem tudor protein ERI-5 potentiates ERI endo-RNAi by tethering an RNA-dependent RNA polymerase (RdRP) module to DCR-1. In the absence of ERI-5, the RdRP module is uncoupled from DCR-1. Notably, EKL-1, an ERI-5 paralog that specifies distinct RdRP modules in Dicer-independent endo-RNAi pathways, partially compensates for the loss of ERI-5 without interacting with DCR-1. Our results implicate tudor proteins in the recruitment of RdRP complexes to specific steps within DCR-1-dependent and DCR-1-independent endo-RNAi pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
09 January 2012
In the version of this article initially published, information in Table 1 was inaccurate. “Newly described” should have been “novel” and “Argonaute protein domain” should have read “Argonaute protein.” The errors have been corrected in the HTML and PDF versions of the article.
References
Mello, C.C. & Conte, D. Jr. Revealing the world of RNA interference. Nature 431, 338–342 (2004).
Tabara, H., Yigit, E., Siomi, H. & Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861–871 (2002).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Yigit, E. et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757 (2006).
Pak, J. & Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244 (2007).
Sijen, T., Steiner, F.A., Thijssen, K.L. & Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007).
Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K. & Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019 (2007).
Guang, S. et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465, 1097–1101 (2010).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Ketting, R.F. The many faces of RNAi. Dev. Cell 20, 148–161 (2011).
Talsky, K.B. & Collins, K. Initiation by a eukaryotic RNA-dependent RNA polymerase requires looping of the template end and is influenced by the template-tailing activity of an associated uridyltransferase. J. Biol. Chem. 285, 27614–27623 (2010).
Claycomb, J.M. et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134 (2009).
Duchaine, T.F. et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354 (2006).
Gu, W. et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244 (2009).
Rocheleau, C.E. et al. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics 178, 1431–1443 (2008).
Conine, C.C. et al. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107, 3588–3593 (2010).
Pavelec, D.M., Lachowiec, J., Duchaine, T.F., Smith, H.E. & Kennedy, S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183, 1283–1295 (2009).
Batista, P.J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Ruby, J.G. et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 (2006).
Vasale, J.J. et al. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107, 3582–3587 (2010).
Gent, J.I. et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37, 679–689 (2010).
Welker, N.C. et al. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893–903 (2010).
Wolters, D.A., Washburn, M.P. & Yates, J.R. III. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 (2001).
Gu, S.G. et al. Distinct ribonucleoprotein reservoirs for microRNA and siRNA populations in C. elegans. RNA 13, 1492–1504 (2007).
Liu, Q. et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301, 1921–1925 (2003).
Han, T. et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106, 18674–18679 (2009).
Colmenares, S.U., Buker, S.M., Buhler, M., Dlakic, M. & Moazed, D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell 27, 449–461 (2007).
Lee, S.R. & Collins, K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 14, 604–610 (2007).
Welker, N.C. et al. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell 41, 589–599 (2011).
Ma, E., MacRae, I.J., Kirsch, J.F. & Doudna, J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380, 237–243 (2008).
Maurer-Stroh, S. et al. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 (2003).
Thomson, T. & Lasko, P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40, 164–170 (2004).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Timmons, L., Court, D.L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001).
Wu, E. et al. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40, 558–570 (2010).
Raymond, C.K., Roberts, B.S., Garrett-Engele, P., Lim, L.P. & Johnson, J.M. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737–1744 (2005).
Acknowledgements
We thank C. Rocheleau for comments on the manuscript, N. Uetani for conceptual and artistic contributions to the model, S. Mitani and his group at the Department of Physiology, Tokyo Women's Medical University School of Medicine, for the generation of the eri-5(tm2528) allele introduced in this manuscript. We thank I. MacRae and N. Welker for discussions on the ERIC model. We also thank A. Haggarty for her assistance in the development of some of the polyclonal antisera. This work was supported by the National Sciences and Engineering Council of Canada RGPIN 341457 (T.F.D.), the Canadian Institute of Health Research MOP 86577 (T.F.D.), the Canada Foundation for Innovation (C.F.I.), and the Fonds de la Rercherche en Santé du Québec, Chercheur-Boursier Salary Award J2 (T.F.D.).
Author information
Authors and Affiliations
Contributions
C.T. conducted the experiments presented in Figures 1c, 2c,d, 3a,b, 4a,b,d and 5a, prepared the figures and assisted with the preparation of the manuscript. N.M. conducted the experiments presented in Figure 1a,d, the ERI-5 samples in 1e and 2a,b. M.F. conducted the experiments presented in Figure 4b,d, and assisted with the model. J.W. carried out the MuDPIT analyses of IP samples. D.C. and J.J.V. conducted the experiments in Figure 3c, under C.C.M.'s direction. D.C. provided scientific advice, and assisted with the redaction of the manuscript. T.F.D. conducted the experiments in Figure 1b, wrote the manuscript and directed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Results and Supplementary Methods (PDF 4186 kb)
Supplementary Data 1
Supplementary data of the coding loci targeted by 26G-RNAs in eri-5 and rrf-3 embryos. (XLSX 117 kb)
Supplementary Data 2
Complement to Supplementary Figure 3: small RNA defect of eri-5 mutant. (XLSX 1556 kb)
Rights and permissions
About this article
Cite this article
Thivierge, C., Makil, N., Flamand, M. et al. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol 19, 90–97 (2012). https://doi.org/10.1038/nsmb.2186
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2186
This article is cited by
-
Asian Citrus Psyllid RNAi Pathway – RNAi evidence
Scientific Reports (2016)
-
Genome wide screening of RNAi factors of Sf21 cells reveal several novel pathway associated proteins
BMC Genomics (2014)
-
Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence
Nature (2013)
-
Erratum: Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi
Nature Structural & Molecular Biology (2013)
-
The molecular architecture of human Dicer
Nature Structural & Molecular Biology (2012)