Abstract
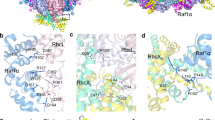
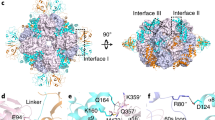
Rubisco, the enzyme that catalyzes the fixation of atmospheric CO2 in photosynthesis, is subject to inactivation by inhibitory sugar phosphates. Here we report the 2.95-Å crystal structure of Nicotiana tabacum Rubisco activase (Rca), the enzyme that facilitates the removal of these inhibitors. Rca from tobacco has a classical AAA+-protein domain architecture. Although Rca populates a range of oligomeric states when in solution, it forms a helical arrangement with six subunits per turn when in the crystal. However, negative-stain electron microscopy of the active mutant R294V suggests that Rca functions as a hexamer. The residues determining species specificity for Rubisco are located in a helical insertion of the C-terminal domain and probably function in conjunction with the N-domain in Rubisco recognition. Loop segments exposed toward the central pore of the hexamer are required for the ATP-dependent remodeling of Rubisco, resulting in the release of inhibitory sugar.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Spreitzer, R.J. & Salvucci, M.E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 (2002).
Andersson, I. & Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 46, 275–291 (2008).
Parry, M.A.J., Keys, A.J., Madgwick, P.J., Carmo-Silva, A.E. & Andralojc, P.J. Rubisco regulation: a role for inhibitors. J. Exp. Bot. 59, 1569–1580 (2008).
Portis, A.R. Jr. Rubisco activase–Rubisco's catalytic chaperone. Photosynth. Res. 75, 11–27 (2003).
Salvucci, M.E. & Crafts-Brandner, S.J. Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol. 134, 1460–1470 (2004).
Kurek, I. et al. Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19, 3230–3241 (2007).
Wang, Z.Y., Snyder, G.W., Esau, B.D., Portis, A.R. Jr. & Ogren, W.L. Species-dependent variation in the interaction of substrate-bound ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and rubisco activase. Plant Physiol. 100, 1858–1862 (1992).
Hanson, P.I. & Whiteheart, S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 (2005).
Li, C., Salvucci, M.E. & Portis, A.R. Two residues of Rubisco activase involved in recognition of the Rubisco substrate. J. Biol. Chem. 280, 24864–24869 (2005).
van de Loo, F.J. & Salvucci, M.E. Activation of ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 35, 8143–8148 (1996).
Esau, B.D., Snyder, G.W. & Portis, A.R. Jr. Differential effects of N- and C-terminal deletions on the two activities of rubisco activase. Arch. Biochem. Biophys. 326, 100–105 (1996).
Roll-Mecak, A. & Vale, R.D. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451, 363–367 (2008).
Guo, F., Maurizi, M.R., Esser, L. & Xia, D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 277, 46743–46752 (2002).
Kim, D.Y. & Kim, K.K. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 278, 50664–50670 (2003).
Blayney, M.J., Whitney, S.M. & Beck, J.L. NanoESI mass spectrometry of Rubisco and Rubisco activase structures and their interactions with nucleotides and sugar phosphates. J. Am. Soc. Mass Spectrom. 22, 1588–1601 (2011).
Li, C., Wang, D. & Portis, A.R. Jr. Identification of critical arginine residues in the functioning of Rubisco activase. Arch. Biochem. Biophys. 450, 176–182 (2006).
Barta, C., Dunkle, A.M., Wachter, R.M. & Salvucci, M.E. Structural changes associated with the acute thermal instability of Rubisco activase. Arch. Biochem. Biophys. 499, 17–25 (2010).
Davies, J.M., Brunger, A.T. & Weis, W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16, 715–726 (2008).
Glynn, S.E., Martin, A., Nager, A.R., Baker, T.A. & Sauer, R.T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009).
Erzberger, J.P. & Berger, J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006).
Mueller-Cajar, O. et al. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature doi:10.1038/nature10568 (2011).
Baker, R.T. et al. Using deubiquitylating enzymes as research tools. Methods Enzymol. 398, 540–554 (2005).
Servaites, J.C. Crystalline ribulose bisphosphate carboxylase/oxygenase of high integrity and catalytic activity from Nicotiana tabacum. Arch. Biochem. Biophys. 238, 154–160 (1985).
Goloubinoff, P., Gatenby, A.A. & Lorimer, G.H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337, 44–47 (1989).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Kreuzer, K.N. & Jongeneel, C.V. Escherichia coli phage T4 topoisomerase. Methods Enzymol. 100, 144–160 (1983).
Barta, C., Carmo-Silva, A.E. & Salvucci, M.E. Rubisco activase activity assays. Methods Mol. Biol. 684, 375–382 (2011).
Smith, J.M. Ximdisp—a visualization tool to aid structure determination from electron microscope images. J. Struct. Biol. 125, 223–228 (1999).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Shaikh, T.R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 (2008).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Pettersen, E.F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994).
Evans, P.R. Scala. in Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography Vol. 33, 22–24 (Daresbury Laboratory, Warrington, UK, 1997).
French, G. & Wilson, K. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Acknowledgements
We thank T. Wauer and L. Popilka for assistance in protein purification and enzyme assays, the Max Planck Institute of Biochemistry Crystallization Facility for support during screening, as well as the Joint Structural Biology Group staff at the European Synchrotron Radiation Facility. Tobacco leaves for the purification of Rubisco were a gift of H.-U. Koob and S. Kirchner (Ludwig-Maximilians-Universität München). The pHUENtRca plasmid was a gift from S. Whitney (Australian National University). We thank the Deutsche Forschungsgemeinschaft (DFG) (SFB 594; DFG grant WE4628/1 to P.W.) and the Körber Foundation for financial support.
Author information
Authors and Affiliations
Contributions
M.S. obtained the Rca crystals and solved the structure together with A.B. M.S. carried out all the biochemical experiments, with help from O.M.-C. The EM and 3D-image analysis were done by S.C. and P.W. All authors contributed to data interpretation and manuscript preparation. M.S., A.B., F.U.H. and M.H.-H. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Table 1 and Supplementary Methods (PDF 1858 kb)
Rights and permissions
About this article
Cite this article
Stotz, M., Mueller-Cajar, O., Ciniawsky, S. et al. Structure of green-type Rubisco activase from tobacco. Nat Struct Mol Biol 18, 1366–1370 (2011). https://doi.org/10.1038/nsmb.2171
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2171
This article is cited by
-
Regulation of Calvin–Benson cycle enzymes under high temperature stress
aBIOTECH (2022)
-
Removal of redox-sensitive Rubisco Activase does not alter Rubisco regulation in soybean
Photosynthesis Research (2022)
-
Decoding the wheat awn transcriptome and overexpressing TaRca1β in rice for heat stress tolerance
Plant Molecular Biology (2021)
-
Enhancing crop yield by using Rubisco activase to improve photosynthesis under elevated temperatures
Stress Biology (2021)
-
Proteome-Wide Analyses Reveal the Diverse Functions of Lysine 2-Hydroxyisobutyrylation in Oryza sativa
Rice (2020)