Abstract
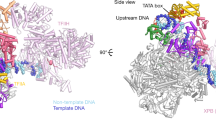
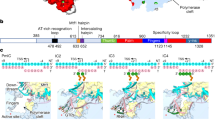
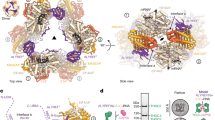
Tfam (transcription factor A, mitochondrial), a DNA-binding protein with tandem high-mobility group (HMG)-box domains, has a central role in the expression, maintenance and organization of the mitochondrial genome. It activates transcription from mitochondrial promoters and organizes the mitochondrial genome into nucleoids. Using X-ray crystallography, we show that human Tfam forces promoter DNA to undergo a U-turn, reversing the direction of the DNA helix. Each HMG-box domain wedges into the DNA minor groove to generate two kinks on one face of the DNA. On the opposite face, a positively charged α-helix serves as a platform to facilitate DNA bending. The structural principles underlying DNA bending converge with those of the unrelated HU family proteins, which have analogous architectural roles in organizing bacterial nucleoids. The functional importance of this extreme DNA bending is promoter specific and seems to be related to the orientation of Tfam on the promoters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Falkenberg, M., Larsson, N.G. & Gustafsson, C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699 (2007).
Bonawitz, N.D., Clayton, D.A. & Shadel, G.S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell 24, 813–825 (2006).
Fisher, R.P. & Clayton, D.A. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8, 3496–3509 (1988).
Fisher, R.P., Topper, J.N. & Clayton, D.A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50, 247–258 (1987).
Larsson, N.G. et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18, 231–236 (1998).
Parisi, M.A. & Clayton, D.A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252, 965–969 (1991).
Stros, M., Launholt, D. & Grasser, K.D. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 64, 2590–2606 (2007).
Dairaghi, D.J., Shadel, G.S. & Clayton, D.A. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 249, 11–28 (1995).
McCulloch, V. & Shadel, G.S. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 23, 5816–5824 (2003).
Kang, D., Kim, S.H. & Hamasaki, N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion 7, 39–44 (2007).
Kaufman, B.A. et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 18, 3225–3236 (2007).
Spelbrink, J.N. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62, 19–32 (2010).
Bogenhagen, D.F., Rousseau, D. & Burke, S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675 (2008).
Alam, T.I. et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31, 1640–1645 (2003).
Ekstrand, M.I. et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13, 935–944 (2004).
Kucej, M., Kucejova, B., Subramanian, R., Chen, X.J. & Butow, R.A. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121, 1861–1868 (2008).
Stott, K., Tang, G.S., Lee, K.B. & Thomas, J.O. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 360, 90–104 (2006).
Fisher, R.P., Parisi, M.A. & Clayton, D.A. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev. 3, 2202–2217 (1989).
Gangelhoff, T.A., Mungalachetty, P.S., Nix, J.C. & Churchill, M.E. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 37, 3153–3164 (2009).
Mouw, K.W. & Rice, P.A. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol. Microbiol. 63, 1319–1330 (2007).
Rice, P.A., Yang, S., Mizuuchi, K. & Nash, H.A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87, 1295–1306 (1996).
Wong, T.S. et al. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 37, 6765–6783 (2009).
Werner, M.H., Huth, J.R., Gronenborn, A.M. & Clore, G.M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell 81, 705–714 (1995).
Kim, J.L., Nikolov, D.B. & Burley, S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365, 520–527 (1993).
Kim, Y., Geiger, J.H., Hahn, S. & Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 365, 512–520 (1993).
Stros, M. HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta 1799, 101–113 (2010).
Ohndorf, U.M., Rould, M.A., He, Q., Pabo, C.O. & Lippard, S.J. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399, 708–712 (1999).
Love, J.J. et al. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376, 791–795 (1995).
Reményi, A. et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 (2003).
Murphy, F.V.IV., Sweet, R.M. & Churchill, M.E. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 18, 6610–6618 (1999).
Luscombe, N.M., Laskowski, R.A. & Thornton, J.M. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 25, 4940–4945 (1997).
Lu, X.J. & Olson, W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003).
Pham, X.H. et al. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 281, 24647–24652 (2006).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Leslie, A.G.W. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography (Warrington WA4 4AD, UK, 1992).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. Data reduction. in Proceedings of the CCP4 Study Weekend. Data Collection and Processing (eds. Sawyer, L., Isaacs, N. & Bailey, S.) 114–122 (Daresbury Laboratory, Warrington, UK, 1993).
Collaborative Computational Project. N. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Painter, J. & Merritt, E.A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 (2006).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Lodeiro, M.F. et al. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J. Biol. Chem. 285, 16387–16402 (2010).
Acknowledgements
We thank N. Chan (California Institute of Technology) for making some mutant constructs, Y. Zhang and Z. Liu (California Institute of Technology) for suggestions on phase determination and structure refinement, T. Walton (California Institute of Technology) for advice on SEC-MALS, S. Shan (California Institute of Technology) for use of equipment and insightful discussions, the staff at the Stanford Synchrotron Radiation Lightsource (SSRL) for technical support with crystal screening and data collection, and members of the Chan laboratory for critical reading of the manuscript. We acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech. SSRL is supported by the US Department of Energy and National Institutes of Health (NIH). This work was supported by NIH grants GM083121 (D.C.C.) and GM062967 (D.C.C.).
Author information
Authors and Affiliations
Contributions
H.B.N. and D.C.C. designed the experiments, analyzed the data and wrote the paper. H.B.N. carried out the crystallography and performed the experimental work. J.T.K. helped with the crystallographic analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Methods (PDF 3672 kb)
Rights and permissions
About this article
Cite this article
Ngo, H., Kaiser, J. & Chan, D. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 18, 1290–1296 (2011). https://doi.org/10.1038/nsmb.2159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2159
This article is cited by
-
Mechanisms and regulation of human mitochondrial transcription
Nature Reviews Molecular Cell Biology (2024)
-
RCCD1 promotes breast carcinogenesis through regulating hypoxia-associated mitochondrial homeostasis
Oncogene (2023)
-
Possible frequent multiple mitochondrial DNA copies in a single nucleoid in HeLa cells
Scientific Reports (2023)
-
The Role of PGC-1α-Mediated Mitochondrial Biogenesis in Neurons
Neurochemical Research (2023)
-
Enhance energy supply of largemouth bass (Micropterus salmoides) in gills during acute hypoxia exposure
Fish Physiology and Biochemistry (2022)