Abstract
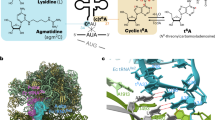
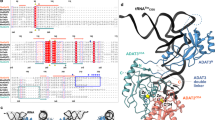
The cytidine at the first position of the anticodon (C34) in the AUA codon-specific archaeal tRNAIle2 is modified to 2-agmatinylcytidine (agm2C or agmatidine), an agmatine-conjugated cytidine derivative, which is crucial for the precise decoding of the genetic code. Agm2C is synthesized by tRNAIle-agm2C synthetase (TiaS) in an ATP-dependent manner. Here we present the crystal structures of the Archaeoglobus fulgidus TiaS–tRNAIle2 complexed with ATP, or with AMPCPP and agmatine, revealing a previously unknown kinase module required for activating C34 by phosphorylation, and showing the molecular mechanism by which TiaS discriminates between tRNAIle2 and tRNAMet. In the TiaS–tRNAIle2–ATP complex, C34 is trapped within a pocket far away from the ATP-binding site. In the agmatine-containing crystals, C34 is located near the AMPCPP γ-phosphate in the kinase module, demonstrating that agmatine is essential for placing C34 in the active site. These observations also provide the structural dynamics for agm2C formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
23 October 2011
In the version of this article initially published online, the last sentence of the abstract was misleading. The error has been corrected for the print, PDF and HTML versions of the article.
References
Björk, G.R. Biosynthesis and function of modified nucleosides. in tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBhandary, U.L.) 165–205 (American Society for Microbiology, Washington, DC, 1995).
Yokoyama, S. & Nishimura, S. Modified nucleosides and codon recognition. in tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBhandary, U.L.) 207–224 (American Society for Microbiology, Washington, DC, 1995).
Suzuki, T. Biosynthesis and function of tRNA wobble modifications. in Topics in Current Genetics: Fine-Tuning of RNA Functions by Modification and Editing (ed. Grosjean, H.) 24–69 (Springer, New York, 2005).
Muramatsu, T. et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181 (1988).
Sylvers, L.A., Rogers, K.C., Shimizu, M., Ohtsuka, E. & Söll, D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 32, 3836–3841 (1993).
Senger, B., Auxilien, S., Englisch, U., Cramer, F. & Fasiolo, F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry 36, 8269–8275 (1997).
Ikeuchi, Y. et al. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6, 277–282 (2010).
Mandal, D. et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNAIle, base pairs with adenosine but not with guanosine. Proc. Natl. Acad. Sci. USA 107, 2872–2877 (2010).
Köhrer, C. et al. Identification and characterization of a tRNA decoding the rare AUA codon in Haloarcula marismortui. RNA 14, 117–126 (2008).
Muramatsu, T. et al. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J. Biol. Chem. 263, 9261–9267 (1988).
Terasaka, N., Kimura, S., Osawa, T., Numata, T. & Suzuki, T. Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS. Nat. Struct. Mol. Biol. 18, doi:10.1038/nsmb.2121 (published online 16 October 2011).
Soma, A. et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell 12, 689–698 (2003).
Ikeuchi, Y. et al. Molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell 19, 235–246 (2005).
Nakanishi, K. et al. Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain. Proc. Natl. Acad. Sci. USA 102, 7487–7492 (2005).
Nakanishi, K. et al. Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase. Nature 461, 1144–1148 (2009).
Kuratani, M. et al. Structural basis of the initial binding of tRNAIle lysidine synthetase TilS with ATP and L-lysine. Structure 15, 1642–1653 (2007).
Suzuki, T. & Miyauchi, K. Discovery and characterization of tRNAIle lysidine synthetase (TilS). FEBS Lett. 584, 272–277 (2010).
Bork, P. & Koonin, E.V. A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins 20, 347–355 (1994).
Carter, A.P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000).
Schmitt, E. et al. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J. 17, 5227–5237 (1998).
Krishna, S.S., Majumdar, I. & Grishin, N.V. Structural classification of zinc fingers. Nucleic Acids Res. 31, 532–550 (2003).
Shi, H. & Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 (2000).
Cusack, S., Yaremchuk, A., Krikliviy, I. & Tukalo, M. tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure 6, 101–108 (1998).
Silvian, L.F., Wang, J. & Steitz, T.A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 (1999).
Eiler, S., Dock-Bregeon, A., Moulinier, L., Thierry, J.C. & Moras, D. Synthesis of aspartyl-tRNAAsp in Escherichia coli–a snapshot of the second step. EMBO J. 18, 6532–6541 (1999).
Fukai, S. et al. Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. RNA 9, 100–111 (2003).
Nakanishi, K., Ogiso, Y., Nakama, T., Fukai, S. & Nureki, O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat. Struct. Mol. Biol. 12, 931–932 (2005).
Hoang, C. & Ferre-D'Amare, A.R. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107, 929–939 (2001).
Lee, T.T., Agarwalla, S. & Stroud, R.M. A unique RNA fold in the RumA–RNA–cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell 120, 599–611 (2005).
Numata, T., Ikeuchi, Y., Fukai, S., Suzuki, T. & Nureki, O. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 442, 419–424 (2006).
Fukuda, W., Morimoto, N., Imanaka, T. & Fujiwara, S. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287, 113–120 (2008).
Morimoto, N. et al. Dual biosynthesis pathway for longer-chain polyamines in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 192, 4991–5001 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Weeks, C.M. & Miller, R. The design and implementation of SnB version 2.0. J. Appl. Crystallogr. 32, 120–124 (1999).
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Abrahams, J.P. & Leslie, A.G.W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 (1996).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Vagin, A. & Teplyakov, A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 (2000).
Acknowledgements
We thank K. Tomita, K. Ito and S. Fukai for valuable and critical comments and suggestions for this manuscript. We thank the beamline staff at BL-17A of the Photon Factory for technical assistance during data collection. This work was supported by a Precursory Research for Embryonic Science and Technology (PRESTO) Program grant from the Japan Science and Technology Agency (JST) (to T.N.); by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (JSPS) (to T.N.); by a Grant-in-Aid for JSPS Fellows (to T.O. and S.K.); by grants from the Kurata Memorial Hitachi Science and Technology Foundation, the Nakajima Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Sankyo Foundation of Life Science and the Noda Institute for Scientific Research (to T.N.); by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.S.); and by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.S.).
Author information
Authors and Affiliations
Contributions
H.I. and T.N. prepared the tRNAIle2 for crystallography. H.I. constructed the protein expression system. T.O. carried out the protein purification, crystallization and structure determination. T.N. assisted with the structure determination. T.O. and H.I. prepared the TiaS mutants. S.K. and N.T. prepared the tRNA mutants. S.K. and N.T. conducted the biochemical analyses under the supervision of T.S., and T.O. and T.N. wrote the paper. All authors discussed the results and commented on the manuscript. T.N. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1 and 2, Supplementary Methods and Supplementary Discussion (PDF 5053 kb)
Rights and permissions
About this article
Cite this article
Osawa, T., Kimura, S., Terasaka, N. et al. Structural basis of tRNA agmatinylation essential for AUA codon decoding. Nat Struct Mol Biol 18, 1275–1280 (2011). https://doi.org/10.1038/nsmb.2144
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2144
This article is cited by
-
Maintenance of the Neuroprotective Function of the Amino Group Blocked Fluorescence-Agmatine
Neurochemical Research (2021)
-
Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS
Nature Structural & Molecular Biology (2011)