Abstract
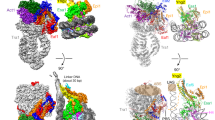
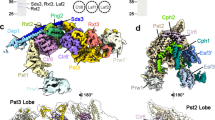
We have used EM and biochemistry to characterize the structure of NuA4, an essential yeast histone acetyltransferase (HAT) complex conserved throughout eukaryotes, and we have determined the interaction of NuA4 with the nucleosome core particle (NCP). The ATM-related Tra1 subunit, which is shared with the SAGA coactivator complex, forms a large domain joined to a second region that accommodates the catalytic subcomplex Piccolo and other NuA4 subunits. EM analysis of a NuA4–NCP complex shows the NCP bound at the periphery of NuA4. EM characterization of Piccolo and Piccolo–NCP provided further information about subunit organization and confirmed that histone acetylation requires minimal contact with the NCP. A small conserved region at the N terminus of Piccolo subunit enhancer of Polycomb-like 1 (Epl1) is essential for NCP interaction, whereas the subunit yeast homolog of mammalian Ing1 2 (Yng2) apparently positions Piccolo for efficient acetylation of histone H4 or histone H2A tails. Taken together, these results provide an understanding of the NuA4 subunit organization and the NuA4–NCP interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Allard, S. et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18, 5108–5119 (1999).
Clarke, A.S., Lowell, J.E., Jacobson, S.J. & Pillus, L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19, 2515–2526 (1999).
Selleck, W., Fortin, I., Sermwittayawong, D., Cote, J. & Tan, S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 25, 5535–5542 (2005).
Boudreault, A.A. et al. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17, 1415–1428 (2003).
Avvakumov, N. & Cote, J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26, 5395–5407 (2007).
Fazzio, T.G., Huff, J.T. & Panning, B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162–174 (2008).
Kim, J. et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 (2010).
Sapountzi, V. & Cote, J. MYST-family histone acetyltransferases: beyond chromatin. Cell. Mol. Life Sci. 68, 1147–1156 (2011).
Doyon, Y. & Cote, J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14, 147–154 (2004).
Auger, A. et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 28, 2257–2270 (2008).
Brown, C.E. et al. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337 (2001).
Downs, J.A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004).
Altaf, M., Auger, A., Covic, M. & Cote, J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell Biol. 87, 35–50 (2009).
Mitchell, L. et al. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 28, 2244–2256 (2008).
Ginsburg, D.S., Govind, C.K. & Hinnebusch, A.G. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29, 6473–6487 (2009).
Carey, M., Li, B. & Workman, J.L. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24, 481–487 (2006).
Altaf, M. et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 285, 15966–15977 (2010).
Grant, P.A. et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650 (1997).
Robert, F. et al. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16, 199–209 (2004).
Pascual-Garcíia, P. et al. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 22, 2811–2822 (2008).
Govind, C.K., Zhang, F., Qiu, H., Hofmeyer, K. & Hinnebusch, A.G. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42 (2007).
Hassan, A.H., Neely, K.E. & Workman, J.L. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104, 817–827 (2001).
Hassan, A.H. et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369–379 (2002).
Yan, Y., Barlev, N.A., Haley, R.H., Berger, S.L. & Marmorstein, R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6, 1195–1205 (2000).
Wang, A.Y. et al. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. USA 106, 21573–21578 (2009).
Sun, B. et al. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J. Biol. Chem. 283, 36504–36512 (2008).
Xu, C., Cui, G., Botuyan, M.V. & Mer, G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16, 1740–1750 (2008).
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001).
Radermacher, M., Wagenknecht, T., Verschoor, A. & Frank, J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146, 113–136 (1987).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
Saxton, W.O. & Baumeister, W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127, 127–138 (1982).
Knutson, B.A. & Hahn, S. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 31, 818–831 (2011).
Wu, P.Y., Ruhlmann, C., Winston, F. & Schultz, P. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15, 199–208 (2004).
Ackerson, C.J., Jadzinsky, P.D., Sexton, J.Z., Bushnell, D.A. & Kornberg, R.D. Synthesis and bioconjugation of 2 and 3 nm-diameter gold nanoparticles. Bioconjug. Chem. 21, 214–218 (2010).
Ackerson, C.J., Jadzinsky, P.D., Jensen, G.J. & Kornberg, R.D. Rigid, specific, and discrete gold nanoparticle/antibody conjugates. J. Am. Chem. Soc. 128, 2635–2640 (2006).
Flaus, A. & Richmond, T.J. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275, 427–441 (1998).
Luger, K., Rechsteiner, T.J. & Richmond, T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999).
Davey, C.A., Sargent, D.F., Luger, K., Maeder, A.W. & Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 (2002).
Berndsen, C.E. et al. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry 46, 2091–2099 (2007).
Stankunas, K. et al. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 125, 4055–4066 (1998).
Nourani, A. et al. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21, 7629–7640 (2001).
Chaban, Y. et al. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat. Struct. Mol. Biol. 15, 1272–1277 (2008).
Carrozza, M.J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007).
Frank, J. Classification of macromolecular assemblies studied as 'single particles'. Q. Rev. Biophys. 23, 281–329 (1990).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Voss, N.R., Yoshioka, C.K., Radermacher, M., Potter, C.S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Daugherty, M.A. & Fried, M.G. Analysis of transcription factor interactions at sedimentation equilibrium. Methods Enzymol. 370, 349–369 (2003).
Cote, J., Utley, R.T. & Workman, J.L. Basic analysis of transcription factor binding to nucleosomes. Methods Mol. Genet. 6, 108–128 (1995).
Acknowledgements
This work was supported by US National Institutes of Health grants R01 GM67167 (F.J.A.), R01 GM060489 (S.T.) and R01 GM070662 (M.G.F.), and fellowship F31 GM086978 (J.R.C.), by a Canadian Institutes of Health Research grant MOP-14308 (J.C.) and fellowship (V.S.), and by a Canada Research Chair (J.C.). We also acknowledge the National Resource for Automated Macromolecular Microscopy.
Author information
Authors and Affiliations
Contributions
M.J.C. and J.M.-S. expressed and purified NuA4 for EM analysis; J.M.-S. expressed and purified the SAGA complex; J.M.-S., R.T.U., V.S., M.C. and S.A. expressed and purified NuA4 and recombinant component subunits for biochemical analysis and performed all biochemical analyses of NuA4 and component subunits; W.S. produced the recombinant NCP; W.S. and J.H. expressed and purified all Piccolo–NuA4 variants and Piccolo–NuA4 in complex with the NCP; M.G.F. performed all analytical ultracentrifugation experiments; Y.C. collected EM data for NuA4 preserved in ice and calculated the RCT 3D reconstruction of NuA4 from stained particles; J.R.C. collected and analyzed all additional EM data and calculated all remaining structures for Piccolo–NuA4 and NuA4; G.C. performed initial quality inspection of 3D NuA4 structures with J.R.C. and F.J.A.; J.R.C., F.J.A., S.T. and J.C. discussed and interpreted all results; F.J.A. supervised all EM-based structural analysis; J.R.C. and F.J.A. wrote the manuscript with input from J.C. and S.T.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 1329 kb)
Rights and permissions
About this article
Cite this article
Chittuluru, J., Chaban, Y., Monnet-Saksouk, J. et al. Structure and nucleosome interaction of the yeast NuA4 and Piccolo–NuA4 histone acetyltransferase complexes. Nat Struct Mol Biol 18, 1196–1203 (2011). https://doi.org/10.1038/nsmb.2128
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2128
This article is cited by
-
A crucial role for dynamic expression of components encoding the negative arm of the circadian clock
Nature Communications (2023)
-
H4 acetylation by the NuA4 complex is required for plastid transcription and chloroplast biogenesis
Nature Plants (2022)
-
Structure of the NuA4 acetyltransferase complex bound to the nucleosome
Nature (2022)
-
Fng1 is involved in crosstalk between histone acetylation and methylation
Current Genetics (2021)
-
Dinucleosome specificity and allosteric switch of the ISW1a ATP-dependent chromatin remodeler in transcription regulation
Nature Communications (2020)