Abstract
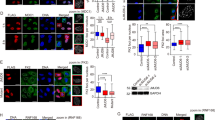
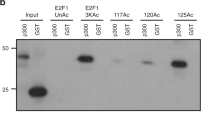
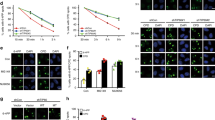
Protein ubiquitination is a crucial component of the DNA damage response. To study the mechanism of the DNA damage–induced ubiquitination pathway, we analyzed the impact of the loss of two E3 ubiquitin ligases, RNF8 and Chfr. Notably, DNA damage–induced activation of ATM kinase is suppressed in cells deficient in both RNF8 and Chfr (double-knockout, or DKO), and DKO mice develop thymic lymphomas that are nearly diploid but harbor clonal chromosome translocations. Moreover, DKO mice and cells are hypersensitive to ionizing radiation. We present evidence that RNF8 and Chfr synergistically regulate histone ubiquitination to control histone H4 Lys16 acetylation through MRG15-dependent acetyltransferase complexes. Through these complexes, RNF8 and Chfr affect chromatin relaxation and modulate ATM activation and DNA damage response pathways. Collectively, our findings demonstrate that two chromatin-remodeling factors, RNF8 and Chfr, function together to activate ATM and maintain genomic stability in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Rouse, J. & Jackson, S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551 (2002).
Harper, J.W. & Elledge, S.J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Khanna, K.K., Lavin, M.F., Jackson, S.P. & Mulhern, T.D. ATM, a central controller of cellular responses to DNA damage. Cell Death Differ. 8, 1052–1065 (2001).
Rotman, G. & Shiloh, Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene 18, 6135–6144 (1999).
Harrison, J.C. & Haber, J.E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 (2006).
Lavin, M.F. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell. Biol. 9, 759–769 (2008).
Barlow, C. et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86, 159–171 (1996).
Xu, Y. et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411–2422 (1996).
Taylor, A.M., Metcalfe, J.A., Thick, J. & Mak, Y.F. Leukemia and lymphoma in ataxia telangiectasia. Blood 87, 423–438 (1996).
Liyanage, M. et al. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood 96, 1940–1946 (2000).
Panier, S. & Durocher, D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst.) 8, 436–443 (2009).
Bennett, E.J. & Harper, J.W. DNA damage: ubiquitin marks the spot. Nat. Struct. Mol. Biol. 15, 20–22 (2008).
Huen, M.S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Kolas, N.K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Wang, B. & Elledge, S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA 104, 20759–20763 (2007).
Ito, K. et al. N-terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur. J. Biochem. 268, 2725–2732 (2001).
Zhao, G.Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007).
Brooks, L. III, Heimsath, E.G. Jr., Loring, G.L. & Brenner, C. FHA-RING ubiquitin ligases in cell division cycle control. Cell. Mol. Life Sci. 65, 3458–3466 (2008).
Bothos, J., Summers, M.K., Venere, M., Scolnick, D.M. & Halazonetis, T.D. The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene 22, 7101–7107 (2003).
Mizuno, K. et al. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene 21, 2328–2333 (2002).
Toyota, M. et al. Epigenetic inactivation of CHFR in human tumors. Proc. Natl. Acad. Sci. USA 100, 7818–7823 (2003).
Huen, M.S. et al. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 28, 6104–6112 (2008).
Bakkenist, C.J. & Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 (2003).
Bertholon, J. et al. Chfr inactivation is not associated to chromosomal instability in colon cancers. Oncogene 22, 8956–8960 (2003).
Lefrançois, D., Kokalj, N., Viegas-Pequignot, E., Montagnier, L. & Dutrillaux, B. High recurrence of rearrangements involving chromosome 14 in an ataxia telangiectasia lymphoblastoid cell line and in its mutagen-treated derivatives. Hum. Genet. 86, 475–480 (1991).
Davey, M.P. et al. Juxtaposition of the T-cell receptor alpha-chain locus (14q11) and a region (14q32) of potential importance in leukemogenesis by a 14;14 translocation in a patient with T-cell chronic lymphocytic leukemia and ataxia-telangiectasia. Proc. Natl. Acad. Sci. USA 85, 9287–9291 (1988).
Baer, R. et al. The breakpoint of an inversion of chromosome 14 in a T-cell leukemia: sequences downstream of the immunoglobulin heavy chain locus are implicated in tumorigenesis. Proc. Natl. Acad. Sci. USA 84, 9069–9073 (1987).
Li, L. et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 207, 983–997 (2010).
Santos, M.A. et al. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J. Exp. Med. 207, 973–981 (2010).
Wu, J. et al. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29, 849–860 (2009).
Lu, L.Y. et al. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 18, 371–384 (2010).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003).
Gordon, F., Luger, K. & Hansen, J.C. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 280, 33701–33706 (2005).
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Rea, S., Xouri, G. & Akhtar, A. Males absent on the first (MOF): from flies to humans. Oncogene 26, 5385–5394 (2007).
Kusch, T. et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 (2004).
Lee, K.K. & Workman, J.L. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 8, 284–295 (2007).
Cai, Y. et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278, 42733–42736 (2003).
Garcia, S.N., Kirtane, B.M., Podlutsky, A.J., Pereira-Smith, O.M. & Tominaga, K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 581, 5275–5281 (2007).
Pardo, P.S., Leung, J.K., Lucchesi, J.C. & Pereira-Smith, O.M. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 277, 50860–50866 (2002).
Prag, G. et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113, 609–620 (2003).
Mueller, T.D. & Feigon, J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 319, 1243–1255 (2002).
Zgheib, O. et al. ATM signaling and 53BP1. Radiother. Oncol. 76, 119–122 (2005).
Bassing, C.H., Swat, W. & Alt, F.W. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109 (Suppl), S45–S55 (2002).
Shiotani, B. & Zou, L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell 33, 547–558 (2009).
You, Z., Bailis, J.M., Johnson, S.A., Dilworth, S.M. & Hunter, T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 9, 1311–1318 (2007).
Kim, Y.C. et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol. 11, 92–96 (2009).
Murr, R. et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91–99 (2006).
Ikura, T. et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27, 7028–7040 (2007).
Sun, Y., Jiang, X., Chen, S., Fernandes, N. & Price, B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 102, 13182–13187 (2005).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Fraga, M.F. et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 (2005).
Sun, Y. et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11, 1376–1382 (2009).
Yu, X. et al. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 37, 401–406 (2005).
Ziv, Y. et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 (2006).
Acknowledgements
We thank E. Fearon, K. Cho, B. Margolis, D. Yali and L. Yang at the University of Michigan for sharing experimental equipment, Z. You at Washington University and K. Tominaga at University of Texas Health Science Center at San Antonio for reagents and J. Keller for proofreading the manuscript. This work was supported by the American Cancer Society (RSG-08-125-01-CCG to X.Y.) and the US National Institutes of Health (CA132755 and CA130899 to X.Y.). X.Y. is a recipient of the Era of Hope Scholar Award from the US Department of Defense.
Author information
Authors and Affiliations
Contributions
J.W. performed most experiments. Y.C. analyzed the MRG15-related protein-protein interactions. L.-Y.L., M.T.P., M.L., Y.W. and D.O.F. provided technical support for various assays. X.Y. designed the experiments. X.Y. and J.W. wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1-7, Supplementary Table 1 and Supplementary Methods (PDF 2455 kb)
Rights and permissions
About this article
Cite this article
Wu, J., Chen, Y., Lu, LY. et al. Chfr and RNF8 synergistically regulate ATM activation. Nat Struct Mol Biol 18, 761–768 (2011). https://doi.org/10.1038/nsmb.2078
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2078
This article is cited by
-
Structural and functional insights into the epigenetic regulator MRG15
Acta Pharmacologica Sinica (2024)
-
DNA damage response revisited: the p53 family and its regulators provide endless cancer therapy opportunities
Experimental & Molecular Medicine (2022)
-
CHFR regulates chemoresistance in triple-negative breast cancer through destabilizing ZEB1
Cell Death & Disease (2021)
-
A synergetic effect of BARD1 mutations on tumorigenesis
Nature Communications (2021)
-
Regulation of DNA damage-induced ATM activation by histone modifications
Genome Instability & Disease (2020)