Abstract
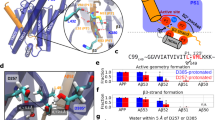
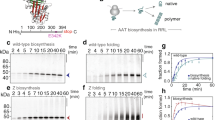
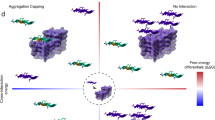
The conformational plasticity of serine protease inhibitors (serpins) underlies both their activities as protease inhibitors and their susceptibility to pathogenic misfolding and aggregation. Here, we structurally characterize a sheet-opened state of the serpin α-1 antitrypsin (α1AT) and show how local unfolding allows functionally essential strand insertion. Mutations in α1AT that cause polymerization-induced serpinopathies map to the labile region, suggesting that the evolution of serpin function required sampling of high risk conformations on a dynamic energy landscape.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gettins, P.G. Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 (2002).
Huntington, J.A., Read, R.J. & Carrell, R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 407, 923–926 (2000).
Dupont, D.M. et al. Biochemical properties of plasminogen activator inhibitor-1. Front. Biosci. 14, 1337–1361 (2009).
Mushunje, A., Evans, G., Brennan, S.O., Carrell, R.W. & Zhou, A. Latent antithrombin and its detection, formation and turnover in the circulation. J. Thromb. Haemost. 2, 2170–2177 (2004).
Kaslik, G. et al. Effects of serpin binding on the target proteinase: global stabilization, localized increased structural flexibility, and conserved hydrogen bonding at the active site. Biochemistry 36, 5455–5464 (1997).
Wang, Z., Mottonen, J. & Goldsmith, E.J. Kinetically controlled folding of the serpin plasminogen activator inhibitor 1. Biochemistry 35, 16443–16448 (1996).
Gooptu, B. & Lomas, D.A. Conformational pathology of the serpins: themes, variations, and therapeutic strategies. Annu. Rev. Biochem. 78, 147–176 (2009).
Tran, S.T. & Shrake, A. The folding of α-1-proteinase inhibitor: kinetic vs equilibrium control. Arch. Biochem. Biophys. 385, 322–331 (2001).
Yamasaki, M., Li, W., Johnson, D.J. & Huntington, J.A. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature 455, 1255–1258 (2008).
Yamasaki, M., Sendall, T.J., Harris, L.E., Lewis, G.M. & Huntington, J.A. The loop-sheet mechanism of serpin polymerization tested by reactive center loop mutations. J. Biol. Chem. 285, 30752–30758 (2010).
Ekeowa, U.I. et al. Defining the mechanism of polymerization in the serpinopathies. Proc. Natl. Acad. Sci. USA 107, 17146–17151 (2010).
James, E.L., Whisstock, J.C., Gore, M.G. & Bottomley, S.P. Probing the unfolding pathway of α1-antitrypsin. J. Biol. Chem. 274, 9482–9488 (1999).
Cabrita, L.D., Whisstock, J.C. & Bottomley, S.P. Probing the role of the F-helix in serpin stability through a single tryptophan substitution. Biochemistry 41, 4575–4581 (2002).
Whisstock, J.C. & Bottomley, S.P. Serpins' mystery solved. Nature 455, 1189–1190 (2008).
Tsutsui, Y. & Wintrode, P.L. Cooperative unfolding of a metastable serpin to a molten globule suggests a link between functional and folding energy landscapes. J. Mol. Biol. 371, 245–255 (2007).
Seo, E.J., Im, H., Maeng, J.S., Kim, K.E. & Yu, M.H. Distribution of the native strain in human α 1-antitrypsin and its association with protease inhibitor function. J. Biol. Chem. 275, 16904–16909 (2000).
Lu, J. & Deutsch, C. PEGylation: a method for assessing topological accessibilities in Kv1.3. Biochemistry 40, 13288–13301 (2001).
Bottomley, S.P., Hopkins, P.C. & Whisstock, J.C. Alpha 1-antitrypsin polymerisation can occur by both loop A and C sheet mechanisms. Biochem. Biophys. Res. Commun. 251, 1–5 (1998).
Tew, D.J. & Bottomley, S.P. Probing the equilibrium denaturation of the serpin α (1)-antitrypsin with single tryptophan mutants; evidence for structure in the urea unfolded state. J. Mol. Biol. 313, 1161–1169 (2001).
Powell, L.M. & Pain, R.H. Effects of glycosylation on the folding and stability of human, recombinant and cleaved α 1-antitrypsin. J. Mol. Biol. 224, 241–252 (1992).
Knaupp, A.S., Levina, V., Robertson, A.L., Pearce, M.C. & Bottomley, S.P. Kinetic instability of the serpin Z α1-antitrypsin promotes aggregation. J. Mol. Biol. 396, 375–383 (2010).
Brantly, M., Courtney, M. & Crystal, R.G. Repair of the secretion defect in the Z form of α 1-antitrypsin by addition of a second mutation. Science 242, 1700–1702 (1988).
Zhou, A., Stein, P.E., Huntington, J.A. & Carrell, R.W. Serpin polymerization is prevented by a hydrogen bond network that is centered on His-334 and stabilized by glycerol. J. Biol. Chem. 278, 15116–15122 (2003).
Kim, J., Lee, K.N., Yi, G.S. & Yu, M.H. A thermostable mutation located at the hydrophobic core of α 1-antitrypsin suppresses the folding defect of the Z-type variant. J. Biol. Chem. 270, 8597–8601 (1995).
Zaimidou, S. et al. A1ATVar: a relational database of human SERPINA1 gene variants leading to α1-antitrypsin deficiency and application of the VariVis software. Hum. Mutat. 30, 308–313 (2009).
Levina, V. et al. Expression, purification and characterization of recombinant Z α (1)-antitrypsin–the most common cause of α (1)-antitrypsin deficiency. Protein Expr. Purif. 68, 226–232 (2009).
Clark, P. & Chong, A.Y. Rare α 1 antitrypsin allele PI W and a history of infant liver disease. Am. J. Med. Genet. 45, 674–676 (1993).
Yu, M.H., Lee, K.N. & Kim, J. The Z type variation of human α 1-antitrypsin causes a protein folding defect. Nat. Struct. Biol. 2, 363–367 (1995).
Lomas, D.A. New insights into the structural basis of α 1-antitrypsin deficiency. Q. J. Med. 89, 807–812 (1996).
Mahadeva, R. et al. Polymers of Z α1-antitrypsin co-localize with neutrophils in emphysematous alveoli and are chemotactic in vivo. Am. J. Pathol. 166, 377–386 (2005).
Elliott, P.R., Pei, X.Y., Dafforn, T.R. & Lomas, D.A. Topography of a 2.0 Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 9, 1274–1281 (2000).
Laska, M.E. The effect of dissolved oxygen on recombinant protein degradation in Escherichia coli. PhD thesis. (Massachusetts Institute of Technology, Cambridge, Massachusetts, USA, 2001).
Ignatova, Z. & Gierasch, L.M. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. USA 103, 13357–13361 (2006).
Nath, U. & Udgaonkar, J.B. Perturbation of a tertiary hydrogen bond in barstar by mutagenesis of the sole His residue to Gln leads to accumulation of at least one equilibrium folding intermediate. Biochemistry 34, 1702–1713 (1995).
Weissman, J.S. & Kim, P.S. Reexamination of the folding of BPTI: predominance of native intermediates. Science 253, 1386–1393 (1991).
Hansen, R.E. & Winther, J.R. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal. Biochem. 394, 147–158 (2009).
Salsbury, F.R. Jr, Knutson, S.T., Poole, L.B. & Fetrow, J.S. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 17, 299–312 (2008).
Acknowledgements
We thank C.L. Cooney (Massachusetts Institute of Technology) for providing a modified form of plasmid pEAT8 (see Online Methods), originally from M.H. Yu (Korea Institute of Science and Technology). We acknowledge S. Eyles and the University of Massachusetts, Amherst, mass spectrometry facility for ESI-MS results, and we thank E. Clerico, D. Hebert and A. Gershenson for stimulating discussions and critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (OD-00045 to L.M.G.) and the Alpha-1 Foundation (to B.K.).
Author information
Authors and Affiliations
Contributions
B.K. and L.M.G. designed the experiments, B.K. carried out the experiments, B.K. and L.M.G. interpreted results and B.K. and L.M.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–3 (PDF 5828 kb)
Rights and permissions
About this article
Cite this article
Krishnan, B., Gierasch, L. Dynamic local unfolding in the serpin α-1 antitrypsin provides a mechanism for loop insertion and polymerization. Nat Struct Mol Biol 18, 222–226 (2011). https://doi.org/10.1038/nsmb.1976
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1976
This article is cited by
-
Nascent chains can form co-translational folding intermediates that promote post-translational folding outcomes in a disease-causing protein
Nature Communications (2021)
-
Aggregation of M3 (E376D) variant of alpha1- antitrypsin
Scientific Reports (2020)
-
High-resolution ex vivo NMR spectroscopy of human Z α1-antitrypsin
Nature Communications (2020)
-
Reactive centre loop dynamics and serpin specificity
Scientific Reports (2019)
-
Smoothing a rugged protein folding landscape by sequence-based redesign
Scientific Reports (2016)